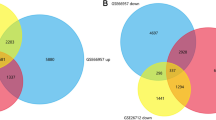
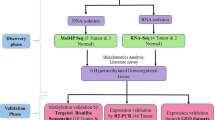
Abstract
Ovarian cancer (OC), one of the most frequent forms of cancer among women all over the world, inflicts a substantial danger to the health of human beings. An in-depth comprehension of its latent processes at the molecular level is the answer to evolving successful earmarked therapies. For an effective genomic assay, deep transcriptional sequencing has been used via Next-generation sequencing tools which are beneficial in studying OC and its related counterparts. ChIP-Seq data for a malignant and benign specimen of OC underwent comparison, and identification of differential peaks were carried out based on fold change and peaks were then annotated. BioGRID was employed to perform protein-protein interaction (PPI) network analysis, which was then constructed with Cytoscape. Highly connected genes from the constructed network were then screened. Utilizing the additional data sets from other OC cell lines gave two new classes of genes for which there is no documented role in the progression of the disease. PAX2, PAX5, FOXP1 and KLF16 are some of the promising genes whose presence among differential peaks led to the positive conclusion of their role in OC. Recent literature studies of these genes are also in conformity with the findings. Potential OC-related genes identified through our findings increase the interpretation of OC and thus provide direction for conducting research in near future.








Similar content being viewed by others
References
Bailey T, Krajewski P, Ladunga I et al (2013) Practical guidelines for the comprehensive analysis of ChIP-seq data. PLoS Comput Biol 9:e1003326
Baylin SB, Ohm JE (2006) Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6:107–116. https://doi.org/10.1038/nrc1799
Bioinformatics B (2011) FastQC: a quality control tool for high throughput sequence data. Camb UK Babraham Inst
Bird AP, Wolffe AP (1999) Methylation-induced repression—belts, braces, and chromatin. Cell 99:451–454. https://doi.org/10.1016/S0092-8674(00)81532-9
Catteau A, Harris WH, Xu C-F, Solomon E (1999) Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene 18:1957–1965. https://doi.org/10.1038/sj.onc.1202509
Chen C, Sun M-Z, Liu S et al (2010) Smad4 mediates malignant behaviors of human ovarian carcinoma cell through the effect on expressions of E-cadherin, plasminogen activator inhibitor-1 and VEGF. Bmb Rep 43:554–560
Choi EJ, Seo EJ, Kim DK et al (2016) FOXP1 functions as an oncogene in promoting cancer stem cell-like characteristics in ovarian cancer cells. Oncotarget 7:3506
Cobaleda C, Schebesta A, Delogu A, Busslinger M (2007) Pax5: the guardian of B cell identity and function. Nat Immunol 8:463
Costello JF, Plass C (2001) Methylation matters. J Med Genet 38:285–303
Darcy KM, Brady WE, Blancato JK et al (2009) Prognostic relevance of c-MYC gene amplification and polysomy for chromosome 8 in suboptimally-resected, advanced stage epithelial ovarian cancers: a gynecologic oncology Group study. Gynecol Oncol 114:472–479. https://doi.org/10.1016/j.ygyno.2009.05.012
Elias KM, Emori MM, Westerling T et al (2016) Epigenetic remodeling regulates transcriptional changes between ovarian cancer and benign precursors. JCI Insight 1
Esteller M (2008) Epigenetics in cancer. N Engl J Med 358:1148–1159. https://doi.org/10.1056/NEJMra072067
Esteller M, Silva JM, Dominguez G et al (2000) Promoter Hypermethylation and BRCA1 Inactivation in Sporadic Breast and Ovarian Tumors. JNCI J Natl Cancer Inst 92:564–569. https://doi.org/10.1093/jnci/92.7.564
Goncharenko-Khaider N, Matte I, Lane D et al (2012) Ovarian cancer ascites increase Mcl-1 expression in tumor cells through ERK1/2-Elk-1 signaling to attenuate TRAIL-induced apoptosis. Mol Cancer 11:84
Hein S, Mahner S, Kanowski C et al (2009) Expression of Jun and Fos proteins in ovarian tumors of different malignant potential and in ovarian cancer cell lines. Oncol Rep 22:177–183
Herman JG, Baylin SB (2003) Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349:2042–2054. https://doi.org/10.1056/NEJMra023075
Imoto I, Sonoda I, Yuki Y, Inazawa J (2001) Identification and characterization of human PKNOX2, a novel homeobox-containing gene. Biochem Biophys Res Commun 287:270–276
Jacobs I, Bast RC (1989) The CA 125 tumour-associated antigen: a review of the literature. Hum Reprod 4:1–12. https://doi.org/10.1093/oxfordjournals.humrep.a136832
Jemal A, Murray T, Ward E et al (2005) Cancer Statistics, 2005. CA Cancer J Clin 55:10–30. https://doi.org/10.3322/canjclin.55.1.10
Johnson DS, Mortazavi A, Myers RM, Wold B (2007) Genome-wide mapping of in vivo protein-DNA interactions. Science 316:1497–1502
Koukoura O, Spandidos DA, Daponte A, Sifakis S (2014) DNA methylation profiles in ovarian cancer: Implication in diagnosis and therapy (Review). Mol Med Rep 10:3–9
Kumar H, Naik PA, Pardasani KR (2017) Finite element model to study calcium distribution in T lymphocyte involving buffers and ryanodine receptors. Proc Natl Acad Sci India Sect Phys Sci. https://doi.org/10.1007/s40010-017-0380-7
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357
Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K (2016) An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol 23:349
Melnikov A, Scholtens D, Godwin A, Levenson V (2009) Differential methylation profile of ovarian cancer in tissues and plasma. J Mol Diagn 11:60–65. https://doi.org/10.2353/jmoldx.2009.080072
Naik PA, Pardasani KR (2015) One dimensional finite element model to study calcium distribution in oocytes in presence of VGCC, RyR and Buffers. http://www.ingentaconnect.com/content/asp/jmihi/2015/00000005/00000003/art00005. Accessed 20 May 2018
Naik PA, Pardasani KR (2016) Finite element model to study calcium distribution in oocytes involving voltage gated Ca2 + channel, ryanodine receptor and buffers. Alex J Med 52:43–49. https://doi.org/10.1016/j.ajme.2015.02.002
Naik PA, Pardasani KR (2018) Three-dimensional finite element model to study effect of RyR calcium channel, ER Leak and SERCA Pump on Calcium Distribution in Oocyte Cell. Int J Comput Methods 1850091
Ortiz L, Aza-Blanc P, Zannini M et al (1999) The interaction between the forkhead thyroid transcription factor TTF-2 and the constitutive factor CTF/NF-1 is required for efficient hormonal regulation of the thyroperoxidase gene transcription. J Biol Chem 274:15213–15221
Raj U, Varadwaj PK (2017) Epigenetics and its role in human cancer. In: Translational bioinformatics and its application. Springer, pp 249–267
Raj U, Kumar H, Gupta S, Varadwaj PK (2016) Exploring dual inhibitors for STAT1 and STAT5 receptors utilizing virtual screening and dynamics simulation validation. J Biomol Struct Dyn 34:2115–2129. https://doi.org/10.1080/07391102.2015.1108870
Raj U, Aier I, Semwal R, Varadwaj PK (2017) Identification of novel dysregulated key genes in breast cancer through high throughput ChIP-Seq data analysis. Sci Rep 7:3229
Scardoni G, Petterlini M, Laudanna C (2009) Analyzing biological network parameters with CentiScaPe. Bioinformatics 25:2857–2859
Semenova EA, Kwon M, Monkhorst K et al (2016) Transcription factor NFIB is a driver of small cell lung cancer progression in mice and marks metastatic disease in patients. Cell Rep 16:631–643
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Song H, Kwan S-Y, Izaguirre DI et al (2013) PAX2 expression in ovarian cancer. Int J Mol Sci 14:6090–6105
Tian C, Ambrosone CB, Darcy KM et al (2012) Common variants in ABCB1, ABCC2 and ABCG2 genes and clinical outcomes among women with advanced stage ovarian cancer treated with platinum and taxane-based chemotherapy: a Gynecologic Oncology Group study. Gynecol Oncol 124:575–581. https://doi.org/10.1016/j.ygyno.2011.11.022
Tuefferd M, Couturier J, Penault-Llorca F et al (2007) HER2 status in ovarian carcinomas: a multicenter GINECO study of 320 patients. Plos One 2:e1138. https://doi.org/10.1371/journal.pone.0001138
Ushijima T, Asada K (2010) Aberrant DNA methylation in contrast with mutations. Cancer Sci 101:300–305. https://doi.org/10.1111/j.1349-7006.2009.01434.x
Wang J, Galvao J, Beach KM et al (2016) Novel roles and mechanism for krüppel-like factor 16 (KLF16) regulation of neurite outgrowth and ephrin receptor A5 (EphA5) expression in retinal ganglion cells. J Biol Chem 291:18084–18095
Weber M, Schübeler D (2007) Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin Cell Biol 19:273–280. https://doi.org/10.1016/j.ceb.2007.04.011
Xia Y, Chang T, Wang Y et al (2014) YAP promotes ovarian cancer cell tumorigenesis and is indicative of a poor prognosis for ovarian cancer patients. PloS One 9:e91770
Xu J, Zhang Y (2012) A generalized linear model for peak calling in ChIP-Seq data. J Comput Biol 19:826–838
Yeung T-L, Leung CS, Wong K-K et al (2017) ELF3 is a negative regulator of epithelial-mesenchymal transition in ovarian cancer cells. Oncotarget 8:16951
Yoo CB, Jones PA (2006) Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov 5:37–50. https://doi.org/10.1038/nrd1930
Zhang Y, Liu T, Meyer CA et al (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137
Acknowledgements
The authors acknowledge the Department of Bioinformatics and Applied Sciences, Indian Institute of Information Technology, Allahabad, for providing computing facility.
Author information
Authors and Affiliations
Contributions
P.K. and U.R. designed computational analyses. P.K. performed the ChIP-Seq data analysis and network studies. P.K., U.R., I.A. and P.K.V. analyzed the data and wrote the paper. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, P., Raj, U., Aier, I. et al. Decoding methylation patterns in ovarian cancer using publicly available Next-Gen sequencing data. Netw Model Anal Health Inform Bioinforma 7, 12 (2018). https://doi.org/10.1007/s13721-018-0173-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-018-0173-1