Abstract
Purpose of Review
The microbiome, defined as the community of microorganisms that live on or in the human body, is involved in a variety of physiological processes. This review summarizes evidence that human microbial communities influence risk of cardiovascular disease (CVD) and place the microbiome in context of other –omic data layers.
Recent Findings
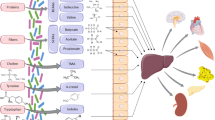
The most robust evidence implicating the microbiome in CVD pathogenesis involves trimethylamine-N-oxide, a moiety synthesized by gut bacteria that has been compellingly linked to the increased risk of adverse CVD events. In addition, many cross-sectional associations have been reported in humans between microbiome composition and various CVD risk factors, including impaired metabolism, hypertension, and inflammation, although interpretation of these correlations is challenging. Host genomic and other –omic variation can mediate associations between microbiome and CVD phenotypes.
Summary
In light of the complexity of –omic data, novel methods are essential to rigorously jointly analyze the role of host and microbial variation in CVD etiology. Until such methods are available, studies would benefit from narrower research questions and a renewed commitment to reproducibility.

Similar content being viewed by others
Change history
28 June 2018
In the recently published paper, “The Interplay Between the Microbiome and Cardiovascular Risk”, the last name of the lead author is listed incorrectly. The author’s name is Brè A. Minniefield.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
World Health Organization. Cardiovascular diseases (CVDs). 2018. http://www.who.int/mediacentre/factsheets/fs317/en/.
Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–322.
•• Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA, et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ res 2015:117:817-824. A large human epidemiologic study of microbiome composition and multiple cardiometabolic phenotypes.
Allayee H, Hazen SL. Contribution of gut bacteria to lipid levels: another metabolic role for microbes? Circ Res. 2015;117:750–4.
Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8:845.
• Brown JM, Hazen SL. Microbial modulation of cardiovascular disease. Nat Rev Microbiol. 2018;16:171–81. A comprehensive review summarizing the mechanisms linking the microbiome (with a focus on the gut) and CVD risk.
Integrative Human Microbiome Project. The integrative human microbiome project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe. 2014;16:276–89.
Clark RI, Walker DW. Role of gut microbiota in aging-related health decline: insights from invertebrate models. Cell Mol Life Sci. 2018;75:93–101.
Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63.
Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–85.
• Heianza Y, Ma W, Manson JE, Rexrode KM, Qi L. Gut microbiota metabolites and risk of major adverse cardiovascular disease events and death: a systematic review and meta-analysis of prospective studies. J Am Heart Assoc. 2017;6:e004947. An up-to-date meta-analysis of gut microbe metabolites (including TMAO) and CVD outcomes.
Tang WHW, Wang Z, Li XS, Fan Y, Li DS, Wu Y, et al. Increased trimethylamine-N-oxide (TMAO) portends high mortality risk independent of glycemic control in patients with type 2 diabetes mellitus. Clin Chem. 2017;63:297–306.
Qi J, You T, Li J, Pan T, Xiang L, Han Y, et al. Circulating trimethylamine-N-oxide and the risk of cardiovascular diseases: a systematic review and meta-analysis of 11 prospective cohort studies. J Cell Mol Med. 2018;22:185–94.
Albert CL, Tang WHW. Metabolic biomarkers in heart failure. Heart Fail Clin. 2018;14:109–18.
Senthong V, Wang Z, Fan Y, Wu Y, Hazen SL, Tang WH. Trimethylamine-N-oxide and mortality risk in patients with peripheral artery disease. J Am Heart Assoc. 2016;5:e004237.
Shafi T, Powe NR, Meyer TW, Hwang S, Hai X, Melamed ML, et al. Trimethylamine-N-oxide and cardiovascular events in hemodialysis patients. J Am Soc Nephrol. 2017;28:321–31.
•• Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–84. The seminal paper linking microbial metabolite TMAO to the risk of CVD.
• Org E, Blum Y, Kasela S, Mehrabian M, Kuusisto J, Kangas AJ, et al. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017;18:70. Cross-sectional large-scale analysis of gut microbiota and metabolic traits, notable for integrating microbiome and metabolome data.
Dalmeijer GW, Olthof MR, Verhoef P, Bots ML, van der Schouw YT. Prospective study on dietary intakes of folate, betaine, and choline and cardiovascular disease risk in women. Eur J Clin Nutr. 2008;62:386–94.
Meyer KA, Shea JW. Dietary choline and betaine and risk of CVD: a systematic review and meta-analysis of prospective studies. Nutrients. 2017;9:e711.
Poesen R, Claes K, Evenepoel P, de Loor H, Augustijns P, Kuypers D, et al. Microbiota-derived phenylacetylglutamine associates with overall mortality and cardiovascular disease in patients with CKD. J Am Soc Nephrol. 2016;27:3479–87.
Ryan PM, Stanton C, Caplice NM. Bile acids at the cross-roads of gut microbiome-host cardiometabolic interactions. Diabetol Metab Syndr. 2017;9:102.
Ranf S, Scheel D, Lee J. Challenges in the identification of microbe-associated molecular patterns in plant and animal innate immunity: a case study with bacterial lipopolysaccharide. Mol Plant Pathol. 2016;17:1165–9.
Haiser HJ, Seim KL, Balskus EP, Turnbaugh PJ. Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes. 2014;5:233–8.
Kholy KE, Genco RJ, Van Dyke TE. Oral infections and cardiovascular disease. Trends Endocrinol Metab. 2015;26:315–21.
Saikku P, Leinonen M, Tenkanen L, Linnanmaki E, Ekman MR, Manninen V, et al. Chronic chlamydia pneumoniae infection as a risk factor for coronary heart disease in the Helsinki heart study. Ann Intern Med. 1992;116:273–8.
Cui L, Zhao T, Hu H, Zhang W, Hua X. Association study of gut flora in coronary heart disease through high-throughput sequencing. Biomed Res Int. 2017;2017:3796359.
Palmer LJ. Complex diseases. In: Elston RC, Olson JM, Palmer LJ, (eds.) Biostatistical genetics and genetic epidemiology. Wiley; 2002.
Kelly TN, Bazzano LA, Ajami NJ, He H, Zhao J, Petrosino JF, et al. Gut microbiome associates with lifetime cardiovascular disease risk profile among Bogalusa heart study participants. Circ Res. 2016;119:956–64.
•• Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–5. The largest to date study of human genomic and phenotypic associations with the gut microbiota, carried out in population-based cohorts.
Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:14.
Yan Q, Gu Y, Li X, Wang W, Jia L, Chen C, et al. Alterations of the gut microbiome in hypertension. Front Cell Infect Microbiol. 2017;7:381.
Kelly CR, Khoruts A, Staley C, Sadowsky MJ, Abd M, Alani M, et al. Effect of fecal microbiota transplantation on recurrence in multiply recurrent Clostridium difficile infection: a randomized trial. Ann Intern Med. 2016;165:609–16.
Al Khodor S, Reichert B, Shatat IF. The microbiome and blood pressure: can microbes regulate our blood pressure? Front Pediatr. 2017;5:138.
Hezel MP, Weitzberg E. The oral microbiome and nitric oxide homoeostasis. Oral Dis. 2015;21:7–16.
Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2010;51:1101–12.
Semova I, Carten JD, Stombaugh J, Mackey LC, Knight R, Farber SA, et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 2012;12:277–88.
Rebolledo C, Cuevas A, Zambrano T, Acuna JJ, Jorquera MA, Saavedra K, et al. Bacterial community profile of the gut microbiota differs between hypercholesterolemic subjects and controls. Biomed Res Int. 2017;6:e005705.
Jones ML, Tomaro-Duchesneau C, Martoni CJ, Prakash S. Cholesterol lowering with bile salt hydrolase-active probiotic bacteria, mechanism of action, clinical evidence, and future direction for heart health applications. Expert Opin Biol Ther. 2013;13:631–42.
Noh DO, Kim SH, Gilliland SE. Incorporation of cholesterol into the cellular membrane of lactobacillus acidophilus ATCC 43121. J Dairy Sci. 1997;80:3107–13.
Veiga P, Juste C, Lepercq P, Saunier K, Beguet F, Gerard P. Correlation between faecal microbial community structure and cholesterol-to-coprostanol conversion in the human gut. FEMS Microbiol Lett. 2005;242:81–6.
Fukushima M, Nakano M. Effects of a mixture of organisms, lactobacillus acidophilus or Streptococcus faecalis on cholesterol metabolism in rats fed on a fat- and cholesterol-enriched diet. Br J Nutr. 1996;76:857–67.
Abdelmaksoud AA, Girerd PH, Garcia EM, Brooks JP, Leftwich LM, Sheth NU, et al. Association between statin use, the vaginal microbiome, and Gardnerella vaginalis vaginolysin-mediated cytotoxicity. PLoS One. 2017;12:e0183765.
Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–77.
Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. 2016;534:213–7.
Kreznar JH, Keller MP, Traeger LL, Rabaglia ME, Schueler KL, Stapleton DS, et al. Host genotype and gut microbiome modulate insulin secretion and diet-induced metabolic phenotypes. Cell Rep. 2017;18:1739–50.
Barlow GM, Yu A, Mathur R. Role of the gut microbiome in obesity and diabetes mellitus. Nutr Clin Pract. 2015;30:787–97.
Kuang YS, Lu JH, Li SH, Li JH, Yuan MY, He JR, et al. Connections between the human gut microbiome and gestational diabetes mellitus. Gigascience. 2017;6:1–12.
Liu F, Li P, Chen M, Luo Y, Prabhakar M, Zheng H, et al. Fructooligosaccharide (FOS) and galactooligosaccharide (GOS) increase Bifidobacterium but reduce butyrate producing bacteria with adverse glycemic metabolism in healthy young population. Sci Rep. 2017;7:11789.
• Buford TW. (Dis)Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome. 2017;5:80. A summary of the microbiome-mediated effects of aging on inflammatory processes, relevant to CVD and other chronic diseases.
Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21:455–66.
Lundberg AM, Hansson GK. Innate immune signals in atherosclerosis. Clin Immunol. 2010;134:5–24.
Brown JM, Hazen SL. The gut microbial endocrine organ: bacterially derived signals driving cardiometabolic diseases. Annu Rev Med. 2015;66:343–59.
Canducci F, Saita D, Foglieni C, Piscopiello MR, Chiesa R, Colombo A, et al. Cross-reacting antibacterial auto-antibodies are produced within coronary atherosclerotic plaques of acute coronary syndrome patients. PLoS One. 2012;7:e42283.
•• Koren O, Spor A, Felin J, Fak F, Stombaugh K, Tremaroli V, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4592–8. Evidence in support of the microbial overlap between the gut, the oral cavity, and atherosclerotic plaques, as well as characterization of both stability and variability of the plaque microbiome.
Chhibber-Goel J, Singhal V, Bhowmik D, Vivek R, Parakh N, Bhargava B, et al. Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. NPJ Biofilms Microbiomes. 2016;2:7.
Lindskog Jonsson A, Hallenius FF, Akrami R, Johansson E, Wester P, Arnerlov C, et al. Bacterial profile in human atherosclerotic plaques. Atherosclerosis. 2017;263:177–83.
De Groot PF, Frissen MN, de Clercq NC, Nieuwdorp M. Fecal microbiota transplantation in metabolic syndrome: history, present and future. Gut Microbes. 2017;8:253–67.
Vrieze A, VanNood E, Holleman F, Salojarvi J, Kootte RS, Barelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–6.
Kootte RS, Levin E, Salojarvi J, Smits LP, Hartstra AV, Udayappan SD, et. al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell 2017:26:611–619.
Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Cultured gut microbiota from twins discordant for obesity modulate adiposity and metabolic phenotypes in mice. Science. 2013;341:10.
Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet. 2017;18:690–9.
•• Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. Robust evidence of interplay across –omic layers.
Goh WWB, Wang W, Wong L. Why batch effects matter in omics data, and how to avoid them. Trends Biotechnol. 2017;35:498–507.
Sharma G, Sowpati DT, Singh P, Khan MZ, Ganji R, Upadhyay S, et al. Genome-wide non-CpG methylation of the host genome during M tuberculosis infection. Sci Rep. 2016;6:25006.
Arora T, Akrami R, Pais R, Bergqvist L, Johansson BR, Schwartz TW, et al. Microbial regulation of the L cell transcriptome. Sci Rep. 2018;8:1207.
Lichtman JS, Alsentzer E, Jaffe M, Sprockett D, Masutani E, Ikwa E, et al. The effect of microbial colonization on the host proteome varies by gastrointestinal location. ISME J. 2016;10:1170–81.
Biteen JS, Blainey PC, Cardon ZG, Chun M, Church GM, Dorrestein PC, et al. Tools for the microbiome: nano and beyond. ACS Nano. 2016;10:6–37.
Chen R, Wu X, Jiang L, Zhang Y. Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep. 2017;18:3227–41.
Shah SH, Newgard CB. Integrated metabolomics and genomics: systems approaches to biomarkers and mechanisms of cardiovascular disease. Circ Cardiovasc Genet. 2015;8:410–9.
Zaiou M, El Amri H. Cardiovascular pharmacogenetics: a promise for genomically-guided therapy and personalized medicine. Clin Genet. 2017;91:355–70.
Ruiz-Canela M, Hruby A, Clish CB, Liang L, Martinez-Gonzalez MA, Hu FB. Comprehensive metabolomic profiling and incident cardiovascular disease: a systematic review. J Am Heart Assoc. 2017;6
Gibbons S, Duvallet C, Alm EJ. Correcting for batch effects in case-control microbiome studies. BioRxiv 2017.
Koleva PT, Bridgman SL, Kozyrskyj AL. The infant gut microbiome: evidence for obesity risk and dietary intervention. Nutrients. 2015;7:2237–60.
González-Reymúndez A, Campos G, Gutiérrez L, Lunt S, Vazquez A. Prediction of years of life after diagnosis of breast cancer using omics and omic-by-treatment interactions. Eur J Hum Genet. 2017;25:538–44.
Hamouda SKM, Abo El-Ezz HR, Wahed ME. Intelligent system for predicting, diagnosis and treatment of breast cancer. Int J Biomed Data Min. 2017;6
Kim D, Joung JG, Sohn KA, Shin H, Park Y, Ritchie M, et al. Knowledge boosting: a graph-based integration approach with multi-omics data and genomic knowledge for cancer clinical outcome prediction. J Am Med Inform Assoc. 2015;22:109–20.
Ebrahim A, Brunk E, Tan J, O’Brien E, Kim D, Szubin R. Multi-omic data integration enables discovery of hidden biological regularities. Nat Commun. 2016;7:13091.
Doostparast TA, Petzold L. Graph-based semi-supervised learning with genomic data integration using condition-responsive genes applied to phenotype classification. J Am Med Inform Assoc. 2018;25:99–108.
Huang S, Chaudhary K, Garmire LX. More is better: recent progress in multi-omics data integration methods. Front Genet. 2017;8:84.
Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:237–45.
Lin D, Zhang J, Li J, Calhoun VD, Deng H-W, Wang Y-P. Group sparse canonical correlation analysis for genomic data integration. BMC Bioinf. 2013;14:245.
Speicher NK, Pfeifer N. Integrating different data types by regularized unsupervised multiple kernel learning with application to cancer subtype discovery. Bioinformatics. 2015;31:268–75.
Fortenberry JD. The uses of race and ethnicity in human microbiome research. Trends Microbiol. 2013;21:165–6.
• Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–8. A classic commentary on the validity of evidence that is applicable to the –omic setting.
Jonsson V, Osterlund T, Nerman O, Kristiansson E. Statistical evaluation of methods for identification of differentially abundant genes in comparative metagenomics. BMC Genomics. 2016;17:78.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Both authors report grants from the National Institutes of Health during the conduct of study.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Cardiovascular Genetics
Rights and permissions
About this article
Cite this article
Minnifield, B.A., Aslibekyan, S.W. The Interplay Between the Microbiome and Cardiovascular Risk. Curr Genet Med Rep 6, 89–97 (2018). https://doi.org/10.1007/s40142-018-0142-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40142-018-0142-0