Abstract
Purpose
Salivary microRNAs (miRNAs) could potentially serve as biomarkers for the diagnosis and prognosis of various types of oral cancer but the significance of it is yet to be fully elucidated. This article reports our study results on the role of salivary miRNAs and their potential use as biomarkers in head and neck cancer (HNC) diagnosis with an emphasis on oral cancers (OC).
Methods
Supernatant saliva samples from 24 subjects, including 12 OC patients and 12 healthy individuals as a control group, were initially profiled using next generation sequencing (NGS). A novel miR-7703 was further validated in 160 samples collected from OC patients (n = 80) and controls (n = 80) by quantitative real-time polymerase chain reaction (qRT-PCR) assays.
Results
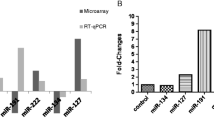
Data analysis revealed that 373 miRNA expressions were significantly decreased and 265 miRNA expressions were significantly increased in patients with OC (p ≤ 0.05). Of the 638 miRNA, twenty-seven were non-redundant miRNAs associated with OC (p ≤ 0.0001). Validation of the novel miR-7703 showed a significant increase in the OC group when compared to the control group. Most importantly, this pattern of increased expression was also positively correlated with tumour stage, lymph node metastasis status, and clinical stage.
Conclusion
This study identified twenty-seven non-redundant, differentially expressed miRNAs associated with OC. These signatures include a number of novel miRNAs as well as those that have been previously reported in either oral or other cancers. However, miR-7703 is a previously uncharacterised miRNA with the potential to be a significant biomarker for the diagnosis of OC.





Similar content being viewed by others
References
Chi AC et al (2009) Differential induction of CYP1A1 and CYP1B1 by benzo[a]pyrene in oral squamous cell carcinoma cell lines and by tobacco smoking in oral mucosa. Oral Oncol 45(11):980–985
Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63(1):11–30
Brinkmann O et al (2011) Oral squamous cell carcinoma detection by salivary biomarkers in a Serbian population. Oral Oncol 47(1):51–55
Patnaik SK et al (2010) Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non–small cell lung cancer after surgical resection. Can Res 70(1):36–45
Calin GA et al (2005) A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353(17):1793–1801
Boeri M et al (2011) MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci 108(9):3713–3718
Fendereski M et al (2017) MicroRNA-196a as a potential diagnostic biomarker for esophageal squamous cell carcinoma. Cancer Invest 35(2):78–84
Baba O et al (2015) MicroRNA-155-5p is associated with oral squamous cell carcinoma metastasis and poor prognosis. J Oral Pathol Med 45:248–255
Christopher AF, Gupta M, Bansal P (2016) Micronome revealed miR-19a/b as key regulator of SOCS3 during cancer related inflammation of oral squamous cell carcinoma. Gene 594(1):30–40
Gorenchtein M et al (2012) MicroRNAs in an oral cancer context—from basic biology to clinical utility. J Dent Res 91(5):440–446
Cervigne NK et al (2009) Identification of a microRNA signature associated with progression of leukoplakia to oral carcinoma. Hum Mol Genet 18(24):4818–4829
Maimaiti A et al (2015) MicroRNA expression profiling and functional annotation analysis of their targets associated with the malignant transformation of oral leukoplakia. Gene 558(2):271–277
Szabo Z et al (2016) Expression of miRNA-21 and miRNA-221 in clear cell renal cell carcinoma (ccRCC) and their possible role in the development of ccRCC. Urol Oncol 34:533-e21
Gorugantula LM et al (2012) Salivary basic fibroblast growth factor in patients with oral squamous cell carcinoma or oral lichen planus. Oral Surg Oral Med Oral Pathol Oral Radiol 114(2):215–222
Cheng YS et al (2011) Salivary endothelin-1 potential for detecting oral cancer in patients with oral lichen planus or oral cancer in remission. Oral Oncol 47(12):1122–1126
Wu B-H et al (2011) MicroRNAs: new actors in the oral cancer scene. Oral Oncol 47(5):314–319
Reis PP et al (2010) Programmed cell death 4 loss increases tumor cell invasion and is regulated by miR-21 in oral squamous cell carcinoma. Mol Cancer 9(1):238
Chen C (2005) MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 353(17):1768
Calin GA et al (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101(9):2999–3004
Manikandan M et al (2015) Down regulation of miR-34a and miR-143 may indirectly inhibit p53 in oral squamous cell carcinoma: a pilot study. Asian Pac J Cancer Prev 16(17):7619–7625
Cummins JM, Velculescu VE (2006) Implications of micro-RNA profiling for cancer diagnosis. Oncogene 25(46):6220–6227
Pal B et al (2015) Integration of microRNA signatures of distinct mammary epithelial cell types with their gene expression and epigenetic portraits. Breast Cancer Res 17:85
Tricoli JV, Jacobson JW (2007) MicroRNA: potential for cancer detection, diagnosis, and prognosis. Cancer Res 67(10):4553–4555
Soga D et al (2013) microRNA expression profiles in oral squamous cell carcinoma. Oncol Rep 30(2):579–583
Liu CJ et al (2012) Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck 34(2):219–224
Park NJ et al (2009) Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res 15(17):5473–5477
Adhami M et al (2017) Candidate miRNAs in human breast cancer biomarkers: a systematic review. Breast Cancer 25:198–205
Momen-Heravi F, Bala S (2018) Emerging role of non-coding RNA in oral cancer. Cell Signal 42:134–143
Liao J et al (2014) Expression profiling of exosomal miRNAs derived from human esophageal cancer cells by Solexa high-throughput sequencing. Int J Mol Sci 15(9):15530–15551
Maltseva DV et al (2014) miRNome of inflammatory breast cancer. BMC Res Notes 7(1):871
Schotte D et al (2011) Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia 25(9):1389–1399
Sand M et al (2016) Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci 83(3):210–218
Sheng Y et al (2014) Downregulation of miR-101-3p by hepatitis B virus promotes proliferation and migration of hepatocellular carcinoma cells by targeting Rab5a. Adv Virol 159(9):2397–2410
Letelier P et al (2014) miR-1 and miR-145 act as tumor suppressor microRNAs in gallbladder cancer. Int J Clin Exp Pathol 7(5):1849–1867
Abdi J et al (2015) Bone marrow stromal cells induce bortezomib resistance in multiple myeloma cells through downregulation of miRNA-101-3p targeting survivin. Blood 126(23):1772
Shaw L, Alder J, Tumilson C (2017) Detection of brain cancer. US Patent 20,170,044,617
Ayaz L et al (2013) Differential expression of microRNAs in plasma of patients with laryngeal squamous cell carcinoma: potential early-detection markers for laryngeal squamous cell carcinoma. J Cancer Res Clin Oncol 139(9):1499–1506
Goto Y et al (2015) Functional significance of aberrantly expressed microRNAs in prostate cancer. Int J Urol 22(3):242–252
Xu H, Liu X, Zhao J (2014) Down-regulation of miR-3928 promoted osteosarcoma growth. Cell Physiol Biochem 33(5):1547–1556
Boeri M et al (2011) MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA 108(9):3713–3718
Aushev VN et al (2013) Comparisons of microRNA patterns in plasma before and after tumor removal reveal new biomarkers of lung squamous cell carcinoma. PLoS ONE 8(10):e78649
Valadi H et al (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9(6):654–659
Acknowledgements
RSF is a recipient of PhD scholarship from Australian Government and Griffith University. RSF wishes to express his gratitude towards the Iraqi Government, Iraqi Center for Cancer and Medical Genetic Research and Al-Mustansiryah University. RSF wishes to thank Dr. David Good, Physiotherapy Department, Australian Catholic University, Queensland, Australia for reading and minor editorial contribution to the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fadhil, R.S., Nair, R.G., Nikolarakos, D. et al. Next generation sequencing identifies novel diagnostic biomarkers for head and neck cancers. Oral Cancer 3, 69–78 (2019). https://doi.org/10.1007/s41548-019-00019-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41548-019-00019-5