Abstract
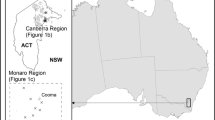
The grassland earless dragon, Tympanocryptis pinguicolla, islisted as endangered throughout its range. A recent taxonomic studybased on both morphological and allozyme data elevatedpinguicolla from a subspecies of T. lineata to fullspecies status, but the allozyme data showed considerabledifferentiation among pinguicolla populations. To investigatethe magnitude and nature of these differences with an independent dataset, we targeted key pinguicolla populations and sequenced anapproximately 900 base pair DNA fragment of the mitochondrial genomethat includes half of the ND4 gene and three tRNA genes. We obtainedsequence data from 21 individuals drawn from the three T.pinguicolla populations, included representatives of two otherTympanocryptis species and used an Amphibolurusspecies as an outgroup. Seven mitochondrial haplotypes were found amongthe 21 T. pinguicolla samples – two in the AustralianCapital Territory (ACT) and five in the Cooma region, Phylogeneticsignal in the data sets was extremely strong and a variety ofphylogenetic analyses of the data all resulted in the same single fullyresolved tree. There are 37 unique differences in the ND4 gene betweenthe ACT and Cooma populations. This translates into genetic differencesof between 5.76% and 6.23% between the two populations. Incomparison to studies on other reptile groups in which the same fragmentof DNA was used, the differences found between the ACT and Coomapopulations are more in line with species-level differences thandifferences within a single species and suggests that these populationsshould be considered separate taxonomic units.
Similar content being viewed by others
References
Arévalo E, Davis SK, Sites J (1994) Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in Central Maxico. Syst. Biol., 43, 387-418.
Benabib M, Kjer KM, Sites JW (1997) Mitochondrial DNA sequence-based phylogeny and the evolution of viviparity in the Sceloporus scalaris group (Reptilia, Squamata). Evolution, 51, 1262-1275.
Brown WM, George M, Wilson AC (1979) Rapid evolution of animal mitochondrial DNA. Proc. Nat. Acad. Sci. USA, 76, 1967-1971.
Cogger HG (2000) Reptiles and Amphibians of Australia, 6th Edition. Reed Books, Chatswood (Australia).
Forstner MRJ, Dixon JR, Forstner JM, Davis SK (1998) Apparent hybridization between Cnemidophorus gularis and Cnemidophorus septemvittatus from an area of sympatry in southwest Texas. J. Herpetology, 32, 418-425.
Hillis D (1991) Discriminating between phylogenetic signal and random noise in DNA sequences. In: Phylogenetic Analysis of DNA Sequences (eds. Miyamoto MM, Cracraft J), pp. 278-294. Oxford University Press, Inc., New York.
Hillis DM, Huelsenbeck JP (1992) Signal, noise, and reliability in molecular phylogenetic analyses. J. Heredity, 83, 189-195.
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol., 42, 182-192.
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Mammalian Protein Metabolism (ed. Munro HN), pp. 21-132. Academic Press, New York.
Kraus F, Mink DG, Brown WM (1996) Crotaline intergeneric relationship based on mitochondrial DNA sequence data. Copeia, 1996, 763-773.
Smith WJS. Osborne WS, Donnellan SC, Cooper PD (1999) The systematic status of earless dragon lizards Tympanocryptis (Reptilia: Agamidae) in south-eastern Australia. Aust. J. Zool., 47, 551-564.
Swofford DL. (2000) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts.
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nuc. Acids Res., 22, 4673-4680.
Wilson AC, Cann RL, Carr SM, George, M Jr., Gyllensten UB, Helm-Bychowski K, Higuchi RC, Palumbi SR, Prager EM, Sage RD, Stoneking M (1985) Mitochondrial DNA and two perspectives on evolutionary genetics. Biol. J. Linn. Soc., 26, 375-400.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Scott, I.A.W., Keogh, J.S. Conservation genetics of the endangered grassland earless dragon Tympanocryptis pinguicolla (Reptilia: Agamidae) in Southeastern Australia. Conservation Genetics 1, 357–363 (2000). https://doi.org/10.1023/A:1011542717349
Issue Date:
DOI: https://doi.org/10.1023/A:1011542717349