Abstract
Background
Myxidium kudoi Meglitsch, 1937 has been described from the type host, blue catfish Ictalurus furcatus, with no additional host records or molecular data available for this species.
Purpose
To provide molecular data and a novel host locality for this species and carry out phylogenetic analyses to infer the evolutionary relationship of the species to other members of the family Myxidiidae for which DNA sequence data is available.
Methods
These data were collected using myxospores from the gallbladder of a blue catfish, Ictalurus furcatus collected from Lake Texoma, Oklahoma, USA. Myxospores were morphologically consistent with the only other account of this species and not any other Myxidium species described from siluriform fishes.
Results
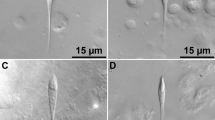
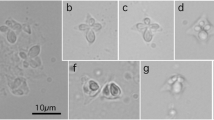
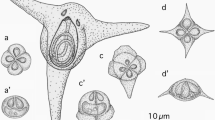
Myxospores were oblong with rounded ends and were 10.8–12.6 (11.6 ± 0.5) µm in length and 4.7–6.6 (5.7 ± 0.5) µm in width. Polar capsules were subspherical and 2.7–3.9 (3.4 ± 0.3) µm in length and 2.4–3.5 (3.1 ± 0.3) µm in diameter, with each capsule containing a polar filament with 3–4 coils. Molecular data consisted of a 2918-bp sequence of the partial 18S, complete ITS1, 5.8S, ITS2, and partial 28S ribosomal rRNA regions as well as a 2455-bp sequence of partial 28S ribosomal RNA. The partial 18S and 28S data was used in a concatenated Bayesian phylogenetic analysis to further infer the evolutionary relationships of the Myxidiidae. Additionally, the partial 18S data was used in a separate phylogenetic analysis.
Conclusions
The present work reports novel morphological and molecular data for Myxidium kudoi as well as a novel locality of occurrence for this species. In concatenated phylogenetic analysis using 18S and 28S data and other molecular data from Myxozoa, M. kudoi grouped with other freshwater Myxidiidae. In the single-locus, 18S analysis, M. kudoi grouped with Myxidium rhodei from Rutilus rutilus and Myxidium amazonense from Corydoras melini, the only other Myxidium species of catfish for which molecular data are available.




Similar content being viewed by others
References
Eiras JC, Saraiva A, Cruz CF, Santos MJ, Fiala I (2011) Synopsis of the species of Myxidium Bütschli, 1882 (Myxozoa: Myxosporea: Bivalvulida). Syst Parasitol 80:81–116. https://doi.org/10.1007/s11230-011-9315-x
Fricke R, Eschmeyer WN, van der Laan WNR (2019) Eschmeyer’s catalog of fishes: genera, species, references. https://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp. Accessed 29 Aug 2019
Meglitsch PA (1937) On some new and known myxosporidia of the fishes of Illinois. J Parasitol 23:467–477. https://doi.org/10.2307/3272393
Fiala I, Bartošová-Sojková P, Whipps CM (2015) Classification and phylogenetics of myxozoa. In: Okamura B, Gruhl A, Bartholomew JL (eds) Myxozoan evolution, ecology and development, 1st edn. Springer, Cham, pp 85–110
Freeman MA, Kristmundson A (2018) Studies of Myxidium giardia Cépède, 1906 infections in Icelandic eels identifies a genetically diverse clade of myxosporeans that represent the Paramyxidium n. g. (Myxosporea: Myxidiidae). Parasit Vectors 11:551. https://doi.org/10.1186/s13071-018-3087-y
Kearse M, Moir R, Wilson A, Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinform 28(12):1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Löytynoja A, Goldman N (2008) A model of evolution and structure for multiple sequence alignment. Philos Trans R Soc B 363:3913–3919. https://doi.org/10.1098/rstb.2008.0170
Misof B, Misof K (2009) A Monte Carlo approach successfully identifies randomness in multiple sequence alingments: a more objective means of data exclusion. Syst Biol 58:21–34
Kück P (2020) Alicut. https://www.zfmk.de/en/research/research-centres-and-groups/utilities. Accessed 13 Jan 2020.
Salinas NR, Little DP (2014) 2matrix: a utility for indel coding and phylogenetic matrix concatenation. Appl Plant Sci 2:1300083. https://doi.org/10.3732/apps.1300083
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49(2):369–381. https://doi.org/10.1093/sysbio/49.2.369
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F (2004) Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407–415. https://doi.org/10.1093/bioinformatics/btg427
Rambaut, A (2016) FigTree. https://tree.bio.ed.ac.uk/software/figtree. Accessed 29 Aug 2019.
Otto GR, Jahn TL (1943) Internal myxosporidian infections of some fishes of the Okoboji region. Proc Iowa Acad Sci 50:323–335
Parker JD, Warner MC (1970) Effects of fixation, dehydration and staining on dimensions of myxosporidian and microsporidian spores. J Wildl Dis 6:448–456. https://doi.org/10.7589/0090-3558-6.4.448
Lom J, Arthur JR (1989) A guideline for the preparation of species descriptions in Myxosporea. J Fish Dis 12:151–156. https://doi.org/10.1111/j.1365-2761.1989.tb00287.x
Molnár K, Ranzani-Paiva MJ, Eiras JC, Rodrigues L (1998) Myxobolus macroplasmodialis sp. n. (Myxozoa: Myxosporea), a parasite of the abdominal cavity of the characid teleost, Salminus maxillosus, in Brazil. Acta Parasitol 37:241–245
Bartošová P, Fiala I, Hypša V (2009) Concatenated SSU and LSU rDNA data confirm the main evolutionary trends within myxosporeans (Myxozoa: Myxosporea) and provide and effective tool for their molecular phylogenetics. Mol Phylogenet Evol 53:81–93. https://doi.org/10.1016/j.ympev.2009.05.018
Mathews PD, Silva MRM, Maia AAM, Adriano EA (2015) Ultrastructure and ssrRNA sequencing of Myxidium amazonense n. sp. a myxosporean parasite of Corydoras melini from the Rio Negro river, Amazonas state Brazil. Parasitol Res 114(12):4675–4683. https://doi.org/10.1007/s00436-015-4715-5
Chenglun M, Xinming D, Cisheng W (1998) The Myxosporidia of freshwater fishes from Sichuan Province I. Myxidium Bütschli and Zschokella Auerbach. J Chongqing Teach Coll (Nat Sci Ed) 15(2):2–10 (In Chinese)
Shulʹman SS (1964) Phylum protozoa. In: Pavlovskii EN (ed) Key to Parasites of freshwater fish of the USSR. Translated from Russian. US Department of the Interior and the National Science Foundation, Washington, pp 5–234
Barta JR, Martin DS, Liberator PA, Dashkevicz M, Anderson JW, Feigner SD, Elbrecht A, Perkins-Barrow A, Jenkins MC, Danforth HD, Ruff MD, Profous-Juchelka H (1997) Phylogenetic relationships among eight Eimeria species infecting domestic fowl inferred using complete small subunit ribosomal DNA sequences. J Parasitol 83:262–271. https://doi.org/10.2307/3284453
Hanson LA, Lin D, Pote LMW, Shivaji R (2001) Small subunit rRNA gene comparisons of four actinosporean species to establish a polymerase chain reaction test for the causative agent of proliferative gill disease in channel catfish. J Aquat Anim Health 13:117–123. https://doi.org/10.1577/1548-8667(2001)013%3c0117:SSRGCO%3e2.0.CO;2
Auwera GV, Chapelle S, Wachter R (1994) megasperma, and phylogeny of the oomycets. FEBS Lett 338:133–136. https://doi.org/10.1016/0014-5793(94)80350-1
Kudo R (1921) On the effects of some fixatives upon myxosporidian spores. Trans Am Microsc Soc 40:161–167
Loch TP, Rosser TG, Baumgartner WA, Boontai T, Faisal M, Griffin MJ (2017) New host record and molecular characterization of Dicauda atherinoidi Hoffman & Walker (Bivalvulida: Myxobolidae): a parasite of the emerald shiner Notropis atherinoides Rafinesque, 1818 and mimic shiner Notropis vollucellus Cope, 1865. J Fish Dis 40:1405–1415
Acknowledgements
A Scientific Collecting Permit was issued to CTM by the Oklahoma Department of Wildlife Conservation (ODWC). CTM thanks the following for assistance in collecting at Lake Texoma: Matt Mauck, David Routledge, and Cliff Sager (ODWC, Caddo, OK, USA) as well as Dr. Don Cloutman (Burdett, KS, USA) for mentoring him on fish myxozoans. The authors acknowledge Wes Baumgartner of Mississippi State University’s College of Veterinary Medicine for histological examination of host tissue samples. This work was supported by the Department of Basic Sciences at Mississippi State University’s College of Veterinary Medicine.
Author information
Authors and Affiliations
Contributions
CTM received and identified the fish host, necropsied the host for parasite collection, and collected and fixed myxospores and infected host tissues. TGR prepared wet mounts of fixed spores, captured photomicrographs, produced plate images, and produced line drawings. ETW measured spores, designed primers used in the present study, carried out DNA extraction and molecular characterisation, compiled relevant literature, and made morphological comparisons with previously described species. TGR and ETW carried out phylogenetic analyses.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author declares that the authors have no conflict of interest affecting this study.
Ethical standards
The fish collected for this study was collected under Oklahoma Department of Wildlife Conservation permit #1551646, issued to CTM. All relevant institutional policies for the ethical use of research animals were followed in handling and processing the fish.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Woodyard, E.T., Rosser, T.G. & McAllister, C.T. Morphological and Molecular Characterisation of Myxidium kudoi Meglitsch, 1937 from the Blue Catfish Ictalurus furcatus, Valenciennes in Oklahoma, USA. Acta Parasit. 65, 388–395 (2020). https://doi.org/10.2478/s11686-020-00172-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11686-020-00172-8