Abstract
Automatic diatom identification approaches have revealed remarkable abilities to tackle the challenges of water quality assessment and other environmental issues. Scientists often analyze the taxonomic characters of the target taxa for automatic identification. In this process the digital photographs, sketches or drawings are recorded to analyze the shape and size of the frustule, the arrangement of striae, the raphe endings, and the striae density. In this paper, we describe two new methods for producing drawings of different diatom species at any stage of their life cycle development that can also be useful for future reference and comparisons. We attempt to produce drawings of diatom species using Edge-preserving Multi-scale Decomposition (EMD). The edge preserving smoothing property of Weighted Least Squares (WLS) optimization framework is used to extract high-frequency details. The details extracted from two-scale decomposition are transformed to drawings which help in identifying possible striae patterns from diatom images. To analyze the salient local features preserved in the drawings, the Scale Invariant Feature Transform (SIFT) model is adopted for feature extraction. The generated drawings help to identify certain unique taxonomic and morphological features that are necessary for the identification of the diatoms. The new methods have been compared with two alternative pencil drawing techniques showing better performance for details preservation.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Diatoms are microscopic microalgae found in aquatic ecosystems that are used as bio-indicators and contribute to the primary production in the aquatic ecosystems. Diatoms have unique taxonomic features called frustule. The main parts of the diatom consist of the nucleus, cell wall, cytoplasm, and plasma membrane. To date, different Light Microscopy (LM) and Scanning Electron Microscopy (SEM) techniques have been developed to visualize the microstructures of the frustule. These taxonomic features are important determinants to classify and identify the diatoms. The automation of classification and identification process plays an important role in a wide range of applications that include forensic examination, ecology, and palaeo-ecology. Nowadays most diatom identification methods demand automatic feature extraction models, which can analyze the photographs and drawings of the diatom species. It is therefore imperative that a method for preparing drawings is needed to record the salient diatom features.
In ADIAC project [5], several attempts to diatom identification and classification were reported. For geometric and texture analysis, different descriptors were utilized that include Scale-invariant Feature Transform (SIFT), Gray-Level Occurrence Matrix (GLCM), Fourier Transform (FT) and Gabor wavelets. It was reported that a 97.97\(\%\) accuracy can be achieved in diatom identification and classification process by using FT and SIFT for the classification of 38 species. Another attempt to explain the diatom classification mechanism is described in DIADIST project [1], in which study on visual indexing of photographs and drawings of microscopic species had been conducted for taxonomic purposes. Extended Depth-of-Focus (DOF) and image-to-drawing conversion methods were applied for taxonomic classification automatically. In a recent study on morphological observation of common pennate diatoms represents that unique features associated with the surface of the frustule are useful for the classification of the diatom [7]. Diatom species were identified based on specific features including the structure of raphe, fibulae and striae, and pore arrangement. Fareha et al. have identified 25 diatom species from the 16 genera from the two estuaries [6].
Drawings have not only morphological or aesthetic interest. They serve to highlight the differential details allowing species discrimination. New diatom taxa need to be accompanied by an illustration showing the morphological differentiation. In recent years there have been some attempts to develop a model for producing drawings of diatom species. McLaughlin has described various methods of drawing generation [14]. The simplest method of creating a drawing is freehand design in which no special apparatus is needed. This process requires observing and recording of fine structures of the frustule manually, which is very time-consuming. Moreover, highly professional and experienced diatomist with a highly developed degree of illustrative ability is required for accurate illustrations. Hicks et al. [12] have proposed an automatic drawing generation model based on frequency and orientation of silica shell patterns, and sternum shapes. Frequency descriptors [20] were utilized to detect the frequency and orientation of silica shell patterns. This model was restricted to the analysis of pennate microscopic species having simple striae patterns on their shells and tested successfully on 12 species. The model was not suitable for species with non-striae patterns, and it seems therefore important to develop automatic drawing generation model that would be adopted for analysis of species with complex and non-striae patterns.
The rest of this paper is organized as follows. The proposed drawing generation models are described in Sect. 2. The experimental results, quantitative and qualitative evaluation, and comparison to other state-of-the-art methods are presented in Sect. 3, and finally, the paper is concluded in Sect. 4.
2 The Proposed Method Using Edge Preserving Filtering
We propose a model for producing drawings of different diatom species based on edge-preserving filter (EPF). We advocate the use of base-detail decomposition technique [8] for multi-scale detail extraction. In many computer graphics and image processing technique, such as HDR tone mapping, detail enhancement [8], image denoising [17] and image fusion [18], it is paramount to operate on images at multiple scales. A number of EPF filter have been proposed [8, 11, 16, 17, 19]. We refer the interested reader to [21] for more comprehensive performance analysis of EPF filters. Among these EPF, the WLS has recently emerged as an excellent tool for multi-scale decomposition, which is suited for single image detail enhancement [8].
WLS Filtering and Base-Detail Layer Decomposition. Prior to generating the drawings of microscopy images of diatom species, we apply multi-scale decomposition, which gives an efficient solution to enhance salient details, while avoiding objectionable artifacts. We would demonstrate that it helps in detecting possible striae patterns from the photographs of diatom species that plays a vital role in producing drawings automatically. In our implementation, let v denote the input microscopy image for which we seek to produce a new image w, which preserve salient edges and contours while smoothing the details between such salient features. For a pixel P, the WLS can be formalized as seeking the minimum of
where \(\gamma \) is a parameter that controls the influence of smoothness weights: increasing the value of \(\gamma \) yields progressively smoother results w. The goal of the expression term \((w_p-v_p)^2\) is to minimize the distance between w and v, while the second (regularization) term strives to achieve smoothness by minimizing the partial derivatives of w. The smoothness requirement is enforced in a spatially varying manner via the smoothness weights \(q_x\) and \(q_y\), which depend on v:
where l is the log-luminance channel of the input image v, the exponent \(\alpha \) determines the sensitivity to the gradients of v, while \(\epsilon \) is a small constant (default value is 0.0001) that avoids division by zero in regions where v is constant.
Using the WLS filter described above, we seek to extract details at multiple scales, and we rely on decomposition similar to Farbman et al. [8]. More specifically, we extract the base layer b and detail layer d from a given input image v. Let \(w^{1},...\,,w^{k}\) represent progressively coarser versions of v. A set of detail layers d are computed as differences between successive coarser levels, and is defined as follows:
In our model the input image is repeatedly smoothed, each time increasing the value of parameter \(\alpha \) by using a fixed value of \(\gamma \) (typically 1.2). Thus, we define:
We have experimented with a six-layer decomposition, in which the initial value of \(\alpha =0.1\) and \(c=2\). Therefore, \(\alpha \) is increased by a factor of 2 at each iteration (i.e. 0.1, 0.2, 0.4, 0.8, 1.6, 3.2). We found that this strategy is suitable to migrate salient features to detail layers for most of the cases while avoiding the effect of noise present in the input images. Several experiments were conducted to decide the free parameters used in WLS filtering. Finally, the detail enhanced image is constructed from the coarsest base layer (i.e. \(b^{6}\)) and six detail layers \(d^{1}, d^{2}, d^{3}, d^{4}, d^{5}, d^{6}\), respectively. To avoid resharpening, sigmoid curve \(S(a,x)=1/\left( 1+exp(-ax)\right) \) is adopted for detail layer manipulation during reconstruction process. More precisely, our detail enhanced image is computed as:
where \(a_{i}\) is the constant depending on subscript \(i=1,...\,,k\). Therefore, \(a_{1}\), \(a_{2}\), \(a_{3}\), \(a_{4}\), \(a_{5}\), \(a_{6}\) are acting as boosting factors for the manipulation of corresponding detail layers (\(d^{1}, d^{2}, d^{3}, d^{4}, d^{5}, d^{6}\)). In our experiments, we have found that \(a_{1}=15\), \(a_{2}=20\), \(a_{3}=80\), \(a_{4}=80\), \(a_{5}=80\), \(a_{6}=80\) are good default setting for most of the cases.
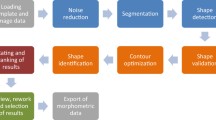
In this paper, we propose two models for generating pencil sketch drawings automatically. The workflow of the first approach called EMD1 is summarized as follows:
-
1.
Load the microscopy image of diatom species.
-
2.
Region of Interest (ROI) selection from input images of diatom species.
-
3.
Construct the detail-enhanced image using Eq. 5.
-
4.
Compute the gradient magnitudes of the image obtained in step 2, using Eq. 6.
-
5.
Construct the negative image of output obtained in step 3, using Eq. 8.
-
6.
Construct the binary image of output obtained in step 4, using Otsu’s method [15].
Region of Selection (ROI) Selection. To select a portion of an image that we want to consider for pencil sketch drawing generation, a binary mask is created. In this binary mask image, pixels that belong to the region of interest (ROI) are set to 1 and all other pixels are set to 0. To select the ROI from microscopy images of diatom species, edge detection and morphological filtering based technique [9] is adopted in this paper. In future work, more sophisticated segmentation methods will be adopted for handling multiple touching objects in the microscopy images of diatom species.
Gradient Computation. In order to detect the striae pattern in an image constructed using Eq. 5, we compute the gradient magnitudes at each point, which is formulated as follows:
where Gx and Gy are the Sobel convolution kernels:
We found that this simple tool is very effective for detecting striae patterns in microscopy images of diatom species.
Inversion and Binarization. To obtain the photographic inverted image of image computed from Eq. 6, we subtract the gradient magnitudes of every pixel from 255:
In the last step, an optimum threshold is computed from gray-level histogram [15] for binarization of drawings. Because of simplicity and suitability, we rely on Otsu’s method that is based on the minimization of inter-class variance. In this approach, the weighted sum of variances of two classes is utilized for computing the optimal threshold T. The final binary image is obtained by applying thresholding operation on image computed from Eq. 8, which is defined as follows:
In the second model of generating pencil sketch drawing called EMD2, all the steps are same except step 3. Instead of computing gradient magnitudes, to detect the image block having salient features such as striae patterns, we apply the filter on \(\hat{w}\) having \(6 \times 6\) kernel:
In the forthcoming section, we will compare the results of proposed models with the existing drawing generation models.
3 Results and Analysis
In this section, we first describe two alternative pencil sketch generation methods available in the literature and then show how proposed multi-scale decomposition helps to produce better results. We also compare our method with recently published work by Hicks et al.
Pencil line drawing is a popular technique to record something that can be utilized as a quick way of graphically signifying a photograph. A pencil drawing also allows an artist to record something to create uncomplicated sketches for later use. In DODGE method [2], the variance of Gaussian was used to blur the inverted grayscale input image. The blend mode technique [3] was used to highlights the salient details such as boldest edges. In another method (2D-CONV) [4], 2D convolution filters were used to transform input photographs into pencil line drawings. Different filter banks were suggested to produce appealing pencil line drawings. Inversion and thresholding operations were used to enhance salient details with some precision to reduce noise. At this point, it is interesting to compare our models to alternative methods. Recall that our objective is to be able to extract shape, size, and pattern of striae on silica shells, so we must keep this objective in mind when analyzing and comparing the alternative methods. Figure 1 demonstrates the results generated from EMD1. The drawings generated from EMD1 preserve the shape, size, and pattern of striae on silica shells. As we mentioned earlier in Sect. 1 that Hicks et al. [12] attempted to address the problem of automatic drawing generation of pennate diatom species having striae pattern. For comparison purpose, we illustrate the drawings of four diatom species in Fig. 2, which are generated automatically through a model proposed by Hicks et al. They also seek objective detecting of striae pattern of the input images, whereas as demonstrated in Fig. 1 we explore the possibilities of extracting an original pattern with shape and size of striae on frustule. The proposed work opens several areas of future research to identify and classify the diatom species from drawings.
Drawings of Hicks et al. [12], generated automatically from 4 diatom species having striae patterns. (a) Caloneis amphisbaena, (b) Cymbella hybrida, (c) Gomphonema augur, and (d) Navicula constans.
Figure 3 shows a comparison of our drawings (see Fig. 3(c, f, i, l)) with the results generated from [4] (see Fig. 3(a, d, g, j)) and [2] (see Fig. 3(b, e, h, k)). A comparison with previous drawing generation models indicates that EMD2 model preserves more salient details from out-of-focus and low contrast regions, as demonstrated in Fig. 3(c, f, i, l). Moreover, EMD2 model provides the control needed to generate more compelling drawings from noisy and low contrast microscopy images of diatom species. Adjusting the free parameter associated with EPF filter in Eq. 5 provides an excellent tool for further interactive adjustments, but for a fair comparison, all the free parameters are kept fixed in this paper.
Comparison of drawings generated from the microscopy images of 4 diatom species: Caloneis amphisbaena (top row, left-to-right), Cymbella hybrida (second row, left-to-right), Gomphonema augur (third row, left-to-right), and Navicula constans (bottom row, left-to-right). (a, d, g, j) 2D-CONV [4], (b, e, h, k) DODGE [2], and (c, f, i, l) Proposed EMD2 model.
To measure the accuracy of proposed EMD2 model, the Scale Invariant Feature Transform (SIFT) model [13] is adopted for feature extraction. The local features extracted from SIFT are highly distinctive and invariant to scale change, affine distortion, change in illumination and addition of noise. [13] demonstrated that invariant local feature matching could be extended to image and pattern recognition problems in which local features were matched against a large number of image data sets. Therefore, to analyze the performance of drawing generation models, we must consider a highly distinctive feature transform such as SIFT. The keypoints extracted using SIFT are shown in Fig. 4. The keypoints extracted from the drawings of [4] are illustrated in Fig. 4(a, d, g, j), and Fig. 4(b, e, h, k) demonstrate the keypoints extracted from the drawings of [2]. In the rightmost drawings (see Fig. 4(c, f, i, l)) of proposed EMD2 model, the goal to preserve more distinctive local features has been achieved. A comparison with previous drawing generation models indicates that EMD2 model preserves more keypoints in the generated drawings.
Visualization of keypoints extracted from drawings of 4 diatom species using SIFT [13]: Caloneis amphisbaena (top row, left-to-right), Cymbella hybrida (second row, left-to-right), Gomphonema augur (third row, left-to-right), and Navicula constans (bottom row, left-to-right). (a, d, g, j) 2D-CONV [4], (b, e, h, k) Dodge [2], and (c, f, i, l) Proposed EMD2 model. The keypoint are extracted from drawings shown in Fig. 3. In order to better visualize the keypoints, a constant value is multiplied to each pixel e.g. 200.
The objective performance analysis of two state-of-the-art pencil line drawing generation methods and proposed models on 4 microscopy images are tabulated in Table 1. In this table, better values are shown in bold (i.e. higher the percentage match of keypoints between input images and drawings is, better the quality of drawings generated). It can be observed from the table that EMD1 has provided better performance for Gomphonema augur and Placoneis constans image data sets. It can be seen that the DODGE outperforms for Caloneis amph. and Cymbella hybrida image data sets but EMD2 models yield the second highest value for Caloneis amph. data set. Similarly, EMD1 models yield the second highest value for Cymbella hybrida data set. Therefore, from the simulation results shown in Table 1, it is clear that among the considered state-of-the-art models, the proposed models have shown better performance.
It is well known that textural feature analysis measures the visual quality of images from different aspects and also useful in image classification problems [10]. In this paper, in addition to visual inspection, to assess the performance of proposed models and other schemes, we consider four textural features computed from grayscale spatial dependencies [10]. In particular, we consider the following Haralick’s co-ocurrence features,
-
Angular Second Moment (ASM), which is a measure of homogeneity of the image.
-
Contrast (C), which is a measure of local intensity variations.
-
Difference Variance (DV).
-
Difference Entropy (DE), which is a measure of information.
For the 4 images of diatom species, comprehensive texture analysis comparisons of proposed models with other schemes are given in Table 2. From Table 2, we can observe that the C, DV, and DE values of EMD1 are always the largest, and the ASM of Caloneis amph. and Cymbella hybrida images are close the highest value. Therefore, the proposed drawing generation models can more effectively extract prominent salient details from input images of diatom species.
4 Conclusions
In this paper, we proposed two models for generating pencil sketch drawings from the images of diatom species. Our proposed models are based on edge-preserving smoothing property of WLS optimization framework. We exhibit the potential of WLS filter for base-detail decomposition, which greatly improves the performance of the proposed models. The experimental results clearly demonstrate that the proposed models can extract the original pattern of stria with shape and size better than other drawing generation models. It should be noted that the WLS fixed parameter set used in multi-scale decomposition can generally obtain good results for most of the images of diatom species. Despite this, we also hypothesize that better results can be generated by choosing different parameters settings according to different types of source images of diatom species, which should be investigated in our future work. In future work, authors would also like to investigate the applicability of proposed models to images of diatom species having non-stria patterns and for other micro-algae organisms.
References
British Diatomists of the 19th Century Database. http://rbg-web2.rbge.org.uk/DIADIST/dia_intro.htm
How to turn any image into a pencil sketch with 10 lines of code. https://bit.ly/2FgkzCV. Accessed 28 Apr 2019
Photoshop Blend Modes Explained. https://bit.ly/2RtJU1z. Accessed 28 Apr 2019
Simple filters and pencil line drawing effect. https://bit.ly/2Vzn1Pg. Accessed 28 Apr 2019
du Buf, H., Bayer, M.M.: Automatic Diatom Identification. Series in Machine Perception and Artificial Intelligence, vol. 51, pp. 1–316. World Scientific, Singapore (2002)
Hilaluddin, F., Leaw, C.P., Lim, P.: Fine structure of the diatoms Thalassiosira and Coscinodiscus (Bacillariophyceae): light and electron microscopy observation. Ann. Microsc. 10, 28–35 (2010)
Hilaluddin, F., Leaw, C.P., Lim, P.: Morphological observation of common pennate diatoms (Bacillariophyceae) from Sarawak estuarine waters. Ann. Microsc. 11, 12–23 (2011)
Farbman, Z., Fattal, R., Lischinski, D., Szeliski, R.: Edge-preserving decompositions for multi-scale tone and detail manipulation. ACM Trans. Graph. 27(3), 67:1–67:10 (2008)
Gonzalez, R.C., Woods, R.E.: Digital Image Processing. Prentice Hall, Upper Saddle River (2008)
Haralick, R.M., Shanmugam, K., Dinstein, I.: Textural features for image classification. IEEE Trans. Syst. Man Cybern. SMC-3(6), 610–621 (1973). https://doi.org/10.1109/TSMC.1973.4309314
He, K., Sun, J., Tang, X.: Guided image filtering. IEEE Trans. Pattern Anal. Mach. Intell. 35(6), 1397–1409 (2013)
Hicks, Y., Marshall, D., Rosin, P.L., Martin, R.R., Mann, D.G., Droop, S.: A model of diatom shape and texture for analysis, synthesis and identification. Mach. Vis. Appl. 17(5), 297–307 (2006)
Lowe, D.G.: Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 60(2), 91–110 (2004)
McLaughlin, R.B.: An Introduction to the Microscopical Study of Diatoms. Hooke College of Applied Sciences, Westmont (2012)
Otsu, N.: A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9(1), 62–66 (1979). https://doi.org/10.1109/TSMC.1979.4310076
Paris, S., Hasinoff, S.W., Kautz, J.: Local Laplacian filters: edge-aware image processing with a Laplacian pyramid. ACM Trans. Graph. 30(4), 68:1–68:12 (2011)
Perona, P., Malik, J.: Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 12(7), 629–639 (1990)
Singh, H., Kumar, V., Bhooshan, S.: Weighted least squares based detail enhanced exposure fusion. ISRN Sig. Process. 2014, 18 (2014)
Tomasi, C., Manduchi, R.: Bilateral filtering for gray and color images. In: Proceedings of the Sixth International Conference on Computer Vision, ICCV 1998. IEEE Computer Society (1998)
Zahn, C.T., Roskies, R.Z.: Fourier descriptors for plane closed curves. IEEE Trans. Comput. C-21(3), 269–281 (1972)
Zhu, F., Liang, Z., Jia, X., Zhang, L., Yu, Y.: A benchmark for edge-preserving image smoothing. IEEE Trans. Image Process. 28, 3556–3570 (2019). A publication of the IEEE Signal Processing Society
Acknowledgements
The authors acknowledge financial support of the Spanish Government under the Aqualitas-retos project (Ref. CTM2014-51907-C2-R-MINECO). The authors thank Saúl Blanco for providing subjective evaluation of the results.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Singh, H., Sánchez, C., Cristóbal, G., Bueno, G. (2019). Pencil Drawing of Microscopic Images Through Edge Preserving Filtering. In: Morales, A., Fierrez, J., Sánchez, J., Ribeiro, B. (eds) Pattern Recognition and Image Analysis. IbPRIA 2019. Lecture Notes in Computer Science(), vol 11868. Springer, Cham. https://doi.org/10.1007/978-3-030-31321-0_17
Download citation
DOI: https://doi.org/10.1007/978-3-030-31321-0_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-31320-3
Online ISBN: 978-3-030-31321-0
eBook Packages: Computer ScienceComputer Science (R0)