Abstract
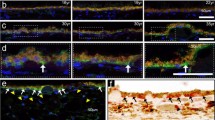
Apolipoprotein D (APOD) is an atypical apolipoprotein with unknown significance for retinal structure and function. Conversely, apolipoprotein E (APOE) is a typical apolipoprotein with established roles in retinal cholesterol transport. Herein, we immunolocalized APOD to the photoreceptor inner segments and conducted ophthalmic characterizations of ApoD−/− and ApoD−/−ApoE−/− mice. ApoD−/− mice had normal levels of retinal sterols but changes in the chorioretinal blood vessels and impaired retinal function. The whole-body glucose disposal was impaired in this genotype but the retinal glucose metabolism was unchanged. ApoD−/−ApoE−/− mice had altered sterol profile in the retina but apparently normal chorioretinal vasculature and function. The whole-body glucose disposal and retinal glucose utilization were enhanced in this genotype. OB-Rb, both leptin and APOD receptor, was found to be expressed in the photoreceptor inner segments and was at increased abundance in the ApoD−/− and ApoD−/−ApoE−/− retinas. Retinal levels of Glut4 and Cd36, the glucose transporter and scavenger receptor, respectively, were increased as well, thus linking APOD to retinal glucose and fatty acid metabolism and suggesting the APOD-OB-Rb-GLUT4/CD36 axis. In vivo isotopic labeling, transmission electron microscopy, and retinal proteomics provided additional insights into the mechanism underlying the retinal phenotypes of ApoD−/− and ApoD−/−ApoE−/− mice. Collectively, our data suggest that the APOD roles in the retina are context specific and could determine retinal glucose fluxes into different pathways. APOD and APOE do not play redundant, complementary or opposing roles in the retina, rather their interplay is more complex and reflects retinal responses elicited by lack of these apolipoproteins.








Similar content being viewed by others
Abbreviations
- APOE:
-
Apolipoprotein E
- APOD:
-
Apolipoprotein D
- AQ4:
-
Aquaporin 4
- ERG:
-
Electroretinographic recordings
- FA:
-
Fluorescein angiography
- FI:
-
Fundus imaging
- GC–MS:
-
Gas chromatography–mass spectroscopy
- HFHS:
-
High-fat high-sugar diet
- IS:
-
Photoreceptor inner segments
- NGS:
-
Normal goat serum
- PBS:
-
Phosphate buffer saline
- RPE:
-
Retinal pigment epithelium
- SD-OCT:
-
Spectral-domain optical coherence tomography
- TCA:
-
The tricarboxylic acid cycle
- TEM:
-
Transmission electron microscopy
References
Fliesler SJ, Bretillon L (2010) The ins and outs of cholesterol in the vertebrate retina. J Lipid Res 51:3399–3413. https://doi.org/10.1194/jlr.R010538
Pikuleva IA, Curcio CA (2014) Cholesterol in the retina: the best is yet to come. Prog Retin Eye Res 41:64–89. https://doi.org/10.1016/j.preteyeres.2014.03.002
Tserentsoodol N, Gordiyenko NV, Pascual I et al (2006) Intraretinal lipid transport is dependent on high density lipoprotein-like particles and class b scavenger receptors. Mol Vis 12:1319–1333
Tserentsoodol N, Sztein J, Campos M et al (2006) Uptake of cholesterol by the retina occurs primarily via a low density lipoprotein receptor-mediated process. Mol Vis 12:1306–1318
Mahley RW (2016) Central nervous system lipoproteins: APOE and regulation of cholesterol metabolism. Arterioscler Thromb Vasc Biol 36:1305–1315. https://doi.org/10.1161/ATVBAHA.116.307023
Saadane A, Petrov A, Mast N et al (2018) Mechanisms that minimize retinal impact of apolipoprotein e absence. J Lipid Res 59:2368–2382. https://doi.org/10.1194/jlr.M090043
Chernick D, Ortiz-Valle S, Jeong A et al (2019) Peripheral versus central nervous system APOE in Alzheimer’s disease: interplay across the blood-brain barrier. Neurosci Lett 708:134306. https://doi.org/10.1016/j.neulet.2019.134306
Kim J, Basak JM, Holtzman DM (2009) The role of apolipoprotein e in Alzheimer’s disease. Neuron 63:287–303. https://doi.org/10.1016/j.neuron.2009.06.026
Zhao N, Liu CC, Qiao W et al (2018) Apolipoprotein e, receptors, and modulation of Alzheimer’s disease. Biol Psychiatry 83:347–357. https://doi.org/10.1016/j.biopsych.2017.03.003
Levy O, Lavalette S, Hu SJ et al (2015) APOE isoforms control pathogenic subretinal inflammation in age-related macular degeneration. J Neurosci 35:13568–13576. https://doi.org/10.1523/jneurosci.2468-15.2015
Mayeux R, Stern Y, Ottman R et al (1993) The apolipoprotein epsilon 4 allele in patients with Alzheimer’s disease. Ann Neurol 34:752–754. https://doi.org/10.1002/ana.410340527
Corder EH, Saunders AM, Strittmatter WJ et al (1993) Gene dose of apolipoprotein e type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261:921–923
Corder EH, Saunders AM, Risch NJ et al (1994) Protective effect of apolipoprotein e type 2 allele for late onset Alzheimer disease. Nat Genet 7:180–184. https://doi.org/10.1038/ng0694-180
Klaver CC, Kliffen M, van Duijn CM et al (1998) Genetic association of apolipoprotein e with age-related macular degeneration. Am J Hum Genet 63:200–206
Souied EH, Benlian P, Amouyel P et al (1998) The epsilon4 allele of the apolipoprotein e gene as a potential protective factor for exudative age-related macular degeneration. Am J Ophthalmol 125:353–359
McKay GJ, Patterson CC, Chakravarthy U et al (2011) Evidence of association of APOE with age-related macular degeneration: a pooled analysis of 15 studies. Hum Mutat 32:1407–1416. https://doi.org/10.1002/humu.21577
Mak AC, Pullinger CR, Tang LF et al (2014) Effects of the absence of apolipoprotein e on lipoproteins, neurocognitive function, and retinal function. JAMA Neurol 71:1228–1236. https://doi.org/10.1001/jamaneurol.2014.2011
Elliott DA, Weickert CS, Garner B (2010) Apolipoproteins in the brain: implications for neurological and psychiatric disorders. Clin Lipidol 51:555–573. https://doi.org/10.2217/CLP.10.37
Drayna D, Fielding C, McLean J et al (1986) Cloning and expression of human apolipoprotein D cDNA. J Biol Chem 261:16535–16539
McConathy WJ, Alaupovic P (1973) Isolation and partial characterization of apolipoprotein D: a new protein moiety of the human plasma lipoprotein system. FEBS Lett 37:178–182. https://doi.org/10.1016/0014-5793(73)80453-3
Perdomo G, Kim DH, Zhang T et al (2010) A role of apolipoprotein d in triglyceride metabolism. J Lipid Res 51:1298–1311. https://doi.org/10.1194/jlr.M001206
Rassart E, Bedirian A, Do Carmo S et al (2000) Apolipoprotein D. Biochim Biophys Acta 1482:185–198
Soiland H, Soreide K, Janssen EA et al (2007) Emerging concepts of apolipoprotein D with possible implications for breast cancer. Cell Oncol 29:195–209
Chen YW, Gregory CM, Scarborough MT et al (2007) Transcriptional pathways associated with skeletal muscle disuse atrophy in humans. Physiol Genomics 31:510–520. https://doi.org/10.1152/physiolgenomics.00115.2006
Dassati S, Waldner A, Schweigreiter R (2014) Apolipoprotein D takes center stage in the stress response of the aging and degenerative brain. Neurobiol Aging 35:1632–1642. https://doi.org/10.1016/j.neurobiolaging.2014.01.148
Eichinger A, Nasreen A, Kim HJ et al (2007) Structural insight into the dual ligand specificity and mode of high density lipoprotein association of apolipoprotein D. J Biol Chem 282:31068–31075. https://doi.org/10.1074/jbc.M703552200
Liu Z, Chang GQ, Leibowitz SF (2001) Apolipoprotein D interacts with the long-form leptin receptor: a hypothalamic function in the control of energy homeostasis. FASEB J 15:1329–1331. https://doi.org/10.1096/fj.00-0530fje
Najyb O, Brissette L, Rassart E (2015) Apolipoprotein D internalization is a basigin-dependent mechanism. J Biol Chem 290:16077–16087. https://doi.org/10.1074/jbc.M115.644302
Lai CJ, Cheng HC, Lin CY et al (2017) Activation of liver x receptor suppresses angiogenesis via induction of ApoD. FASEB J. https://doi.org/10.1096/fj.201700374R
Sarjeant JM, Lawrie A, Kinnear C et al (2003) Apolipoprotein D inhibits platelet-derived growth factor-bb-induced vascular smooth muscle cell proliferated by preventing translocation of phosphorylated extracellular signal regulated kinase 1/2 to the nucleus. Arterioscler Thromb Vasc Biol 23:2172–2177. https://doi.org/10.1161/01.ATV.0000100404.05459.39
D’Souza AM, Neumann UH, Glavas MM et al (2017) The glucoregulatory actions of leptin. Mol Metab 6:1052–1065. https://doi.org/10.1016/j.molmet.2017.04.011
Banks AS, Davis SM, Bates SH et al (2000) Activation of downstream signals by the long form of the leptin receptor. J Biol Chem 275:14563–14572. https://doi.org/10.1074/jbc.275.19.14563
Bahrenberg G, Behrmann I, Barthel A et al (2002) Identification of the critical sequence elements in the cytoplasmic domain of leptin receptor isoforms required for janus kinase/signal transducer and activator of transcription activation by receptor heterodimers. Mol Endocrinol 16:859–872. https://doi.org/10.1210/mend.16.4.0800
Hileman SM, Pierroz DD, Masuzaki H et al (2002) Characterization of short isoforms of the leptin receptor in rat cerebral microvessels and of brain uptake of leptin in mouse models of obesity. Endocrinology 143:775–783. https://doi.org/10.1210/endo.143.3.8669
Blanco-Vaca F, Via DP, Yang CY et al (1992) Characterization of disulfide-linked heterodimers containing apolipoprotein d in human plasma lipoproteins. J Lipid Res 33:1785–1796
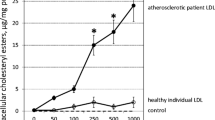
Tsukamoto K, Mani DR, Shi J et al (2013) Identification of apolipoprotein d as a cardioprotective gene using a mouse model of lethal atherosclerotic coronary artery disease. Proc Natl Acad Sci USA 110:17023–17028. https://doi.org/10.1073/pnas.1315986110
Ganfornina MD, Do Carmo S, Lora JM et al (2008) Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell 7:506–515. https://doi.org/10.1111/j.1474-9726.2008.00395.x
Balbin M, Freije JM, Fueyo A et al (1990) Apolipoprotein D is the major protein component in cyst fluid from women with human breast gross cystic disease. Biochem J 271:803–807. https://doi.org/10.1042/bj2710803
Alvarez ML, Barbon JJ, Gonzalez LO et al (2003) Apolipoprotein D expression in retinoblastoma. Ophthalmic Res 35:111–116. https://doi.org/10.1159/000069130
Hunter S, Weiss S, Ou CY et al (2005) Apolipoprotein D is down-regulated during malignant transformation of neurofibromas. Hum Pathol 36:987–993. https://doi.org/10.1016/j.humpath.2005.06.018
Jin D, El-Tanani M, Campbell FC (2006) Identification of apolipoprotein D as a novel inhibitor of osteopontin-induced neoplastic transformation. Int J Oncol 29:1591–1599
Sasaki Y, Negishi H, Koyama R et al (2009) P53 family members regulate the expression of the apolipoprotein D gene. J Biol Chem 284:872–883. https://doi.org/10.1074/jbc.M807185200
Martineau C, Najyb O, Signor C et al (2016) Apolipoprotein D deficiency is associated to high bone turnover, low bone mass and impaired osteoblastic function in aged female mice. Metabolism 65:1247–1258. https://doi.org/10.1016/j.metabol.2016.05.007
Hitman GA, McCarthy MI, Mohan V et al (1992) The genetics of non-insulin-dependent diabetes mellitus in south india: an overview. Ann Med 24:491–497. https://doi.org/10.3109/07853899209167001
Vijayaraghavan S, Hitman GA, Kopelman PG (1994) Apolipoprotein-D polymorphism: a genetic marker for obesity and hyperinsulinemia. J Clin Endocrinol Metab 79:568–570. https://doi.org/10.1210/jcem.79.2.7913935
Baker WA, Hitman GA, Hawrami K et al (1994) Apolipoprotein D gene polymorphism: a new genetic marker for type 2 diabetic subjects in Nauru and South India. Diabet Med 11:947–952. https://doi.org/10.1111/j.1464-5491.1994.tb00252.x
Desai PP, Bunker CH, Ukoli FA et al (2002) Genetic variation in the apolipoprotein D gene among african blacks and its significance in lipid metabolism. Atherosclerosis 163:329–338. https://doi.org/10.1016/s0021-9150(02)00012-6
Desai PP, Hendrie HC, Evans RM et al (2003) Genetic variation in apolipoprotein D affects the risk of Alzheimer disease in African-Americans. Am J Med Genet B Neuropsychiatr Genet 116B:98–101. https://doi.org/10.1002/ajmg.b.10798
Helisalmi S, Hiltunen M, Vepsalainen S et al (2004) Genetic variation in apolipoprotein D and Alzheimer’s disease. J Neurol 251:951–957. https://doi.org/10.1007/s00415-004-0470-8
Waldner A, Dassati S, Redl B et al (2018) Apolipoprotein D concentration in human plasma during aging and in Parkinson’s disease: a cross-sectional study. Parkinsons Dis 2018:3751516. https://doi.org/10.1155/2018/3751516
Loerch PM, Lu T, Dakin KA et al (2008) Evolution of the aging brain transcriptome and synaptic regulation. PLoS ONE 3:e3329. https://doi.org/10.1371/journal.pone.0003329
de Magalhaes JP, Curado J, Church GM (2009) Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25:875–881. https://doi.org/10.1093/bioinformatics/btp073
Lee CK, Weindruch R, Prolla TA (2000) Gene-expression profile of the ageing brain in mice. Nat Genet 25:294–297. https://doi.org/10.1038/77046
Terrisse L, Seguin D, Bertrand P et al (1999) Modulation of apolipoprotein D and apolipoprotein E expression in rat hippocampus after entorhinal cortex lesion. Brain Res Mol Brain Res 70:26–35
Jansen PJ, Lutjohann D, Thelen KM et al (2009) Absence of ApoE upregulates murine brain ApoD and ABCA1 levels, but does not affect brain sterol levels, while human ApoE3 and human ApoE4 upregulate brain cholesterol precursor levels. J Alzheimers Dis 18:319–329. https://doi.org/10.3233/JAD-2009-1150
Levros LC Jr, Labrie M, Charfi C et al (2013) Binding and repressive activities of apolipoprotein E3 and E4 isoforms on the human ApoD promoter. Mol Neurobiol 48:669–680. https://doi.org/10.1007/s12035-013-8456-0
Liu L, MacKenzie KR, Putluri N et al (2017) The glia-neuron lactate shuttle and elevated ROS promote lipid synthesis in neurons and lipid droplet accumulation in glia via APOE/D. Cell Metab 26(719–737):e716. https://doi.org/10.1016/j.cmet.2017.08.024
Kalaany NY, Mangelsdorf DJ (2006) LXRs and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol 68:159–191. https://doi.org/10.1146/annurev.physiol.68.033104.152158
Boyles JK, Notterpek LM, Anderson LJ (1990) Accumulation of apolipoproteins in the regenerating and remyelinating mammalian peripheral nerve. Identification of apolipoprotein D, apolipoprotein A-IV, apolipoprotein E, and apolipoprotein A-I. J Biol Chem 265:17805–17815
Montpied P, de Bock F, Lerner-Natoli M et al (1999) Hippocampal alterations of apolipoprotein E and D mRNA levels in vivo and in vitro following kainate excitotoxicity. Epilepsy Res 35:135–146. https://doi.org/10.1016/s0920-1211(99)00003-0
Kosacka J, Gericke M, Nowicki M et al (2009) Apolipoproteins D and E3 exert neurotrophic and synaptogenic effects in dorsal root ganglion cell cultures. Neuroscience 162:282–291. https://doi.org/10.1016/j.neuroscience.2009.04.073
Do Carmo S, Seguin D, Milne R et al (2002) Modulation of apolipoprotein D and apolipoprotein E mRNA expression by growth arrest and identification of key elements in the promoter. J Biol Chem 277:5514–5523. https://doi.org/10.1074/jbc.M105057200
Schafer NF, Luhmann UF, Feil S et al (2009) Differential gene expression in Ndph-knockout mice in retinal development. Investig Ophthalmol Vis Sci 50:906–916. https://doi.org/10.1167/iovs.08-1731
Zheng W, Mast N, Saadane A et al (2015) Pathways of cholesterol homeostasis in mouse retina responsive to dietary and pharmacologic treatments. J Lipid Res 56:81–97. https://doi.org/10.1194/jlr.M053439
Omarova S, Charvet CD, Reem RE et al (2012) Abnormal vascularization in mouse retina with dysregulated retinal cholesterol homeostasis. J Clin Investig 122:3012–3023. https://doi.org/10.1172/JCI63816
Saadane A, Mast N, Charvet CD et al (2014) Retinal and nonocular abnormalities in Cyp27a1(−/−)Cyp46a1(−/−) mice with dysfunctional metabolism of cholesterol. Am J Pathol 184:2403–2419. https://doi.org/10.1016/j.ajpath.2014.05.024
Curcio CA, Rudolf M, Wang L (2009) Histochemistry and lipid profiling combine for insights into aging and age-related maculopathy. Methods Mol Biol 580:267–281. https://doi.org/10.1007/978-1-60761-325-1_15
Zheng W, Reem RE, Omarova S et al (2012) Spatial distribution of the pathways of cholesterol homeostasis in human retina. PLoS ONE 7:e37926. https://doi.org/10.1371/journal.pone.0037926
Saadane A, Mast N, Trichonas G et al (2019) Retinal vascular abnormalities and microglia activation in mice with deficiency in cytochrome p450 46a1-mediated cholesterol removal. Am J Pathol 189:405–425. https://doi.org/10.1016/j.ajpath.2018.10.013
Mast N, Reem R, Bederman I et al (2011) Cholestenoic acid is an important elimination product of cholesterol in the retina: comparison of retinal cholesterol metabolism with that in the brain. Investig Ophthalmol Vis Sci 52:594–603. https://doi.org/10.1167/iovs.10-6021
Mast N, Shafaati M, Zaman W et al (2010) Marked variability in hepatic expression of cytochromes CYP7A1 and CYP27A1 as compared to cerebral CYP46A1. Lessons from a dietary study with omega 3 fatty acids in hamsters. Biochim Biophys Acta 1801:674–681. https://doi.org/10.1016/j.bbalip.2010.03.005
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Zhang J, Xin L, Shan B et al (2012) Peaks DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics 11(M111):010587. https://doi.org/10.1074/mcp.M111.010587
Karnovsky MJ (1965) A formaldehyde-glutaraldehyde fixative of high osmolarity for use electron microscopy. J Cell Biol 27:137A
van Dijk TH, van der Sluijs FH, Wiegman CH et al (2001) Acute inhibition of hepatic glucose-6-phosphatase does not affect gluconeogenesis but directs gluconeogenic flux toward glycogen in fasted rats. A pharmacological study with the chlorogenic acid derivative S4048. J Biol Chem 276:25727–25735. https://doi.org/10.1074/jbc.M101223200
Pfrieger FW, Ungerer N (2011) Cholesterol metabolism in neurons and astrocytes. Prog Lipid Res 50:357–371. https://doi.org/10.1016/j.plipres.2011.06.002
Mast N, Bederman IR, Pikuleva IA (2018) Retinal cholesterol content is reduced in simvastatin-treated mice due to inhibited local biosynthesis albeit increased uptake of serum cholesterol. Drug Metab Dispos 46:1528–1537. https://doi.org/10.1124/dmd.118.083345
Castanho MA, Coutinho A, Prieto MJ (1992) Absorption and fluorescence spectra of polyene antibiotics in the presence of cholesterol. J Biol Chem 267:204–209
Coscas F, Coscas G, Souied E et al (2007) Optical coherence tomography identification of occult choroidal neovascularization in age-related macular degeneration. Am J Ophthalmol 144:592–599. https://doi.org/10.1016/j.ajo.2007.06.014
Robson JG, Frishman LJ (2014) The rod-driven a-wave of the dark-adapted mammalian electroretinogram. Prog Retin Eye Res 39:1–22. https://doi.org/10.1016/j.preteyeres.2013.12.003
Petrov AM, Astafev AA, Mast N et al (2019) The interplay between retinal pathways of cholesterol output and its effects on mouse retina. Biomolecules. https://doi.org/10.3390/biom9120867
Westermann B (2010) Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11:872–884. https://doi.org/10.1038/nrm3013
Hsieh WT, Hsu CJ, Capraro BR et al (2012) Curvature sorting of peripheral proteins on solid-supported wavy membranes. Langmuir 28:12838–12843. https://doi.org/10.1021/la302205b
Ait-Ali N, Fridlich R, Millet-Puel G et al (2015) Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell 161:817–832. https://doi.org/10.1016/j.cell.2015.03.023
Steinbusch LK, Schwenk RW, Ouwens DM et al (2011) Subcellular trafficking of the substrate transporters GLUT4 and CD36 in cardiomyocytes. Cell Mol Life Sci 68:2525–2538. https://doi.org/10.1007/s00018-011-0690-x
Febbraio M, Hajjar DP, Silverstein RL (2001) CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Investig 108:785–791. https://doi.org/10.1172/jci14006
Kerner J, Hoppel C (2000) Fatty acid import into mitochondria. Biochim Biophys Acta 1486:1–17. https://doi.org/10.1016/s1388-1981(00)00044-5
Tang J, Kern TS (2011) Inflammation in diabetic retinopathy. Prog Retin Eye Res 30:343–358. https://doi.org/10.1016/j.preteyeres.2011.05.002
Durham JT, Herman IM (2011) Microvascular modifications in diabetic retinopathy. Curr Diab Rep 11:253–264. https://doi.org/10.1007/s11892-011-0204-0
Stitt AW, Lois N, Medina RJ et al (2013) Advances in our understanding of diabetic retinopathy. Clin Sci (Lond) 125:1–17. https://doi.org/10.1042/CS20120588
Radu M, Chernoff J (2013) An in vivo assay to test blood vessel permeability. J Vis Exp. https://doi.org/10.3791/50062
Ong JM, Zorapapel NC, Rich KA et al (2001) Effects of cholesterol and apolipoprotein E on retinal abnormalities in ApoE-deficient mice. Investig Ophthalmol Vis Sci 42:1891–1900
Ong JM, Zorapapel NC, Aoki AM et al (2003) Impaired electroretinogram (ERG) response in apolipoprotein E-deficient mice. Curr Eye Res 27:15–24
Dithmar S, Curcio CA, Le NA et al (2000) Ultrastructural changes in Bruch’s membrane of apolipoprotein E-deficient mice. Investig Ophthalmol Vis Sci 41:2035–2042
Kliffen M, Lutgens E, Daemen MJ et al (2000) The APO(*)E3-leiden mouse as an animal model for basal laminar deposit. Br J Ophthalmol 84:1415–1419
Malek G, Johnson LV, Mace BE et al (2005) Apolipoprotein E allele-dependent pathogenesis: a model for age-related retinal degeneration. Proc Natl Acad Sci USA 102:11900–11905. https://doi.org/10.1073/pnas.0503015102
Levy O, Calippe B, Lavalette S et al (2015) Apolipoprotein E promotes subretinal mononuclear phagocyte survival and chronic inflammation in age-related macular degeneration. EMBO Mol Med 7:211–226. https://doi.org/10.15252/emmm.201404524
Maioli S, Lodeiro M, Merino-Serrais P et al (2015) Alterations in brain leptin signalling in spite of unchanged CSF leptin levels in Alzheimer’s disease. Aging Cell 14:122–129. https://doi.org/10.1111/acel.12281
Konstantinidis D, Paletas K, Koliakos G et al (2009) Signaling components involved in leptin-induced amplification of the atherosclerosis-related properties of human monocytes. J Vasc Res 46:199–208. https://doi.org/10.1159/000161234
Wang JL, Chinookoswong N, Scully S et al (1999) Differential effects of leptin in regulation of tissue glucose utilization in vivo. Endocrinology 140:2117–2124. https://doi.org/10.1210/endo.140.5.6681
Sanchez-Chavez G, Pena-Rangel MT, Riesgo-Escovar JR et al (2012) Insulin stimulated-glucose transporter glut 4 is expressed in the retina. PLoS ONE 7:e52959. https://doi.org/10.1371/journal.pone.0052959
Naeser P (1997) Insulin receptors in human ocular tissues. Immunohistochemical demonstration in normal and diabetic eyes. Ups J Med Sci 102:35–40. https://doi.org/10.3109/03009739709178930
Joyal JS, Sun Y, Gantner ML et al (2016) Retinal lipid and glucose metabolism dictates angiogenesis through the lipid sensor Ffar1. Nat Med 22:439–445. https://doi.org/10.1038/nm.4059
Fielding PE, Fielding CJ (1980) A cholesteryl ester transfer complex in human plasma. Proc Natl Acad Sci USA 77:3327–3330. https://doi.org/10.1073/pnas.77.6.3327
Wong-Riley MT (2010) Energy metabolism of the visual system. Eye Brain 2:99–116. https://doi.org/10.2147/EB.S9078
Mantych GJ, Hageman GS, Devaskar SU (1993) Characterization of glucose transporter isoforms in the adult and developing human eye. Endocrinology 133:600–607. https://doi.org/10.1210/endo.133.2.8344201
Watanabe T, Mio Y, Hoshino FB et al (1994) Glut2 expression in the rat retina: localization at the apical ends of muller cells. Brain Res 655:128–134. https://doi.org/10.1016/0006-8993(94)91606-3
Kam JH, Jeffery G (2015) To unite or divide: mitochondrial dynamics in the murine outer retina that preceded age related photoreceptor loss. Oncotarget 6:26690–26701. https://doi.org/10.18632/oncotarget.5614
Poitry-Yamate CL, Poitry S, Tsacopoulos M (1995) Lactate released by muller glial cells is metabolized by photoreceptors from mammalian retina. J Neurosci 15:5179–5191
Winkler BS, Arnold MJ, Brassell MA et al (1997) Glucose dependence of glycolysis, hexose monophosphate shunt activity, energy status, and the polyol pathway in retinas isolated from normal (nondiabetic) rats. Investig Ophthalmol Vis Sci 38:62–71
Bringmann A, Grosche A, Pannicke T et al (2013) GABA and glutamate uptake and metabolism in retinal glial (muller) cells. Front Endocrinol (Lausanne) 4:48. https://doi.org/10.3389/fendo.2013.00048
Thanos S, Bohm MR, Meyer zu Horste M et al (2014) Role of crystallins in ocular neuroprotection and axonal regeneration. Prog Retin Eye Res 42:145–161. https://doi.org/10.1016/j.preteyeres.2014.06.004
Philp NJ, Yoon MY, Hock RS (1990) Identification and localization of talin in chick retinal pigment epithelial cells. Exp Eye Res 51:191–198
Sheedy FJ, Grebe A, Rayner KJ et al (2013) CD36 coordinates NLRP3 inflammasome activation by facilitating intracellular nucleation of soluble ligands into particulate ligands in sterile inflammation. Nat Immunol 14:812–820. https://doi.org/10.1038/ni.2639
Ryeom SW, Silverstein RL, Scotto A et al (1996) Binding of anionic phospholipids to retinal pigment epithelium may be mediated by the scavenger receptor CD36. J Biol Chem 271:20536–20539
Houssier M, Raoul W, Lavalette S et al (2008) CD36 deficiency leads to choroidal involution via COX2 down-regulation in rodents. PLoS Med 5:e39. https://doi.org/10.1371/journal.pmed.0050039
Picard E, Houssier M, Bujold K et al (2010) CD36 plays an important role in the clearance of oxLDL and associated age-dependent sub-retinal deposits. Aging (Albany NY) 2:981–989. https://doi.org/10.18632/aging.100218
Nagelhus EA, Veruki ML, Torp R et al (1998) Aquaporin-4 water channel protein in the rat retina and optic nerve: polarized expression in muller cells and fibrous astrocytes. J Neurosci 18:2506–2519
Kumar B, Gupta SK, Srinivasan BP et al (2013) Hesperetin rescues retinal oxidative stress, neuroinflammation and apoptosis in diabetic rats. Microvasc Res 87:65–74. https://doi.org/10.1016/j.mvr.2013.01.002
Kumar B, Gupta SK, Nag TC et al (2014) Retinal neuroprotective effects of quercetin in streptozotocin-induced diabetic rats. Exp Eye Res 125:193–202. https://doi.org/10.1016/j.exer.2014.06.009
Xu HP, Zhao JW, Yang XL (2002) Expression of voltage-dependent calcium channel subunits in the rat retina. Neurosci Lett 329:297–300. https://doi.org/10.1016/s0304-3940(02)00688-2
Nimmrich V, Gross G (2012) P/Q-type calcium channel modulators. Br J Pharmacol 167:741–759. https://doi.org/10.1111/j.1476-5381.2012.02069.x
Lohner M, Babai N, Muller T et al (2017) Analysis of rim expression and function at mouse photoreceptor ribbon synapses. J Neurosci 37:7848–7863. https://doi.org/10.1523/JNEUROSCI.2795-16.2017
Grabner CP, Gandini MA, Rehak R et al (2015) RIM1/2-mediated facilitation of Cav1.4 channel opening is required for Ca2+-stimulated release in mouse rod photoreceptors. J Neurosci 35:13133–13147. https://doi.org/10.1523/JNEUROSCI.0658-15.2015
Acknowledgements
This work was supported in part by NIH grants EY018383 and EY011373 (I.A.P.) and the unrestricted grant from Research to Prevent Blindness. Irina A. Pikuleva is a Carl F. Asseff Professor of Ophthalmology. The authors thank the Visual Sciences Research Center Core Facilities (supported by National Institutes of Health Grant P30 EY11373) for assistance with mouse breeding (Heather Butler and Kathryn Franke), animal genotyping (John Denker), tissue sectioning (Catherine Doller), and microscopy (Scott Howell and Anthony Gardella). We are also grateful to Dr. Hisashi Fujioka (Electron Microscopy Core facility) for help with studies of retinal ultrastructure, to Danie Schlatzer (Proteomics and Small Molecule Mass Spectrometry Core) for conducting retinal label free analysis, and to Dr. Neal Peachey for assistance with the ERG studies.
Author information
Authors and Affiliations
Contributions
IAP and AMP conception and study design; IAP study supervision; NED, NM, AMP, TD, AAA, AS, EP, ES, and IB produced and analyzed data; IAP, NM, AMP, and IB wrote and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
El-Darzi, N., Mast, N., Petrov, A.M. et al. Studies of ApoD−/− and ApoD−/−ApoE−/− mice uncover the APOD significance for retinal metabolism, function, and status of chorioretinal blood vessels. Cell. Mol. Life Sci. 78, 963–983 (2021). https://doi.org/10.1007/s00018-020-03546-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-020-03546-3