Abstract
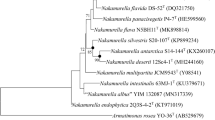
A novel light-yellow-coloured, Gram-stain-positive, nearly-coccoid, aerobic bacterium, designated strain ID2601ST was isolated from a car evaporator core collected from South Korea. Strain ID2601ST was catalase positive and oxidase negative, able to grow at pH 6.0–8.0, temperature 20–45 °C, and 0–6.0% (w/v) NaCl concentration. The 16S rRNA gene sequence analysis showed that strain ID2601ST belonged to the genus Flexivirga, with the nearest phylogenetic neighbour being Flexivirga endophytica YIM 7505T (97.9% sequence similarity). The strain comprised diphosphatidylglycerol as the main polar lipid; MK-8(H4) as a predominant respiratory quinone; serine, alanine, glycine, glutamic acid, and lysine as main components of peptidoglycan and iso-C16:0, summed feature 9 (iso-C17:1ω9c and/or C16:0 10-methyl), anteiso-C17:0, and C17:0 10-methyl as the major fatty acids. The average nucleotide identity (ANI) values between strain ID2601ST and the closest species (Flexivirga endophytica YIM 7505T and Flexivirga caeni BO-16T) were < 78%. The in silico DNA–DNA hybridization (dDDH) values of strain ID2601ST with the closest species were < 22%. These observations were below the threshold values of 95% (for ANI) and 70% (for dDDH) used for species delineation. The DNA G+C content was 69.8 mol%. Based on the polyphasic taxonomic data, the novel species Flexivirga aerilata sp. nov. is proposed with the type strain ID2601ST (=KCTC 49353T =NBRC 114622T).

Similar content being viewed by others
References
Anzai K, Sugiyama T, Sukisaki M et al (2011) Flexivirga alba gen. nov., sp. nov., an actinobacterial taxon in the family Dermacoccaceae. J Antibiot 64:613–616
Keum DH, Lee YJ, Lee SY, Im WT (2020) Flexivirga caeni sp. nov., isolated from activated sludge. Int J Syst Evol Microbiol 70:1266–1272
Gao R, Liu BB, Yang W et al (2016) Flexivirga endophytica sp. nov., an endophytic actinobacterium isolated from a leaf of Sweet Basil. Int J Syst Evol Microbiol 66:3388–3392
Kang W, Hyun DW, Kim PS et al (2016) Flexivirga lutea sp. nov., isolated from the faeces of a crested ibis, Nipponia nippon, and emended description of the genus Flexivirga. Int J Syst Evol Microbiol 66:3594–3599
Hyeon JW, Kim HR, Lee HJ et al (2017) Flexivirga oryzae sp. nov., isolated from soil of a rice paddy, and emended description of the genus Flexivirga Anzai et al. 2012. Int J Syst Evol Microbiol 67:479–484
Frank JA, Reich CI, Sharma S et al (2008) Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 74:2461–2470
Chaudhary DK, Kim J (2018) Flavobacterium naphthae sp. nov., isolated from oil-contaminated soil. Int J Syst Evol Microbiol 68:305–309
Yoon SH, Ha SM, Kwon S et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Pruesse E, Peplies J, Glöckner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N Y) 39:783–791
Coil D, Jospin G, Darling AE (2015) A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Lee I, Chalita M, Ha S-M et al (2017) ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int J Syst Evol Microbiol 67:2053–2057
Tatusova T, DiCuccio M, Badretdin A et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624
Aziz RK, Bartels D, Best AA et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75
Blin K, Shaw S, Steinke K et al (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87
Yoon S-H, Ha S, Lim J et al (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Doetsch RN (1981) Determinative methods of light microscopy. In: Gerhardt P, Murray RGE, Costilow RN et al (eds) Manual of methods for general bacteriology. American Society for Microbiology, Washington DC, pp 21–33
Breznak JA, Costilow RN (2007) Physicochemical factors in growth. In: Reddy CA, Beveridge TJ, Breznak JA et al (eds) Methods for general and molecular bacteriology, 3rd edn. American Society of Microbiology, Washinton, DC, pp 309–329
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Minnikin DE, O’Donnell AG, Goodfellow M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev 45:316–354
Komagata K, Suzuki KI (1988) 4 Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Bousfield GR, Sugino H, Ward DN (1985) Demonstration of a COOH-terminal extension on equine lutropin by means of a common acid-labile bond in equine lutropin and equine chorionic gonadotropin. J Biol Chem 260:9531–9533
Sasser M (1990) Bacterial identification by gas chromatographic analysis of fatty acid methyl esters (GC-FAME). MIDI Tech Note 101 Newwark, MIDI Inc
Kim M, Oh H-S, Park S-C, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Wayne LG, Brenner DJ, Colwell RR et al (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464
Meier-Kolthoff JP, Göker M, Spröer C, Klenk HP (2013) When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol 195:413–418
Acknowledgements
This research was supported by Sangji University Research Fund, 2020 and grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR202002108).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest regarding the publication of this manuscript.
Ethical Approval
This study does not describe any experimental work related to human.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chaudhary, D.K., Lee, H., Dahal, R.H. et al. Flexivirga aerilata sp. nov., Isolated from an Automobile Air Conditioning System. Curr Microbiol 78, 796–802 (2021). https://doi.org/10.1007/s00284-020-02300-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02300-z