Abstract
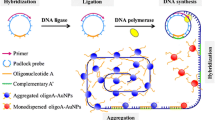
The authors have incidentally found that the three tandem repeats of a 13-mer G-rich oligomer (with sequence 5′-TGG GAA GGG AGG G-3′; referred to as G3) can directly fold into a stable G3 trimer. The G3 trimer/hemin DNAzyme exhibits an about 3-fold higher peroxidase-mimicking activity compared to the conventional G3/hemin DNAzyme. Combining this finding with rolling circle amplification (RCA), a colorimetric assay was developed for sensitive and specific determination of microRNA. In this method, each cycle of RCA generates three catalytic units. This leads to a significant signal amplification of the RCA. Using let-7a as a model analyte, the colorimetric method (best performed at 420 nm) exhibits high sensitivity toward microRNA-let-7a with a 37 fM detection limit and an analytical range that covers 3 orders of magnitude. The method was applied to the determination of let-7a in some cell lysates.

This G-triplex trimer-based rolling circle amplification (RCA) method can produce three catalytic units per RCA cycle, which can significantly improve the amplification efficiency of RCA.






Similar content being viewed by others
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism and function. Cell 116:281–297
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435:828–833
Tricoli JV, Jacobson JW (2007) MicroRNA: potential for cancer detection, diagnosis, and prognosis. Cancer Res 67:4553–4555
Schmittgen TD, Jiang J, Liu Q, Yang L (2004) A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 32:e43
Wang X, Wang H, Liu C, Wang H, Li Z (2017) A three-way junction structure-based isothermal exponential amplification strategy for sensitive detection of 3′-terminal 2'-O-methylated plant microRNA. Chem Commun 53:1124–1127
Qu X, Jin H, Liu Y, Sun Q (2018) Strand displacement amplification reaction on quantum dot-encoded silica bead for visual detection of multiplex microRNAs. Anal Chem 90:3482–3489
Guo J, Mingoes C, Qiu X, Hildebrandt N (2019) Simple, amplified, and multiplexed detection of microRNAs using time-gated FRET and hybridization chain reaction. Anal Chem 91:3101–3109
Yu S, Wang Y, Jiang LP, Bi S, Zhu JJ (2018) Cascade amplification-mediated in situ hot-spot assembly for microRNA detection and molecular logic gate operations. Anal Chem 90:4544–4551
Zhou F, Meng R, Liu Q, Jin Y, Li B (2016) Photoinduced electron transfer-based fluorescence quenching combined with rolling circle amplification for sensitive detection of microRNA. ChemistrySelect 1:6422–6428
Zhou F, Li B, Ma J (2015) A linear DNA probe as an alternative to a molecular beacon for improving the sensitivity of a homogenous fluorescence biosensing platform for DNA detection using target-primed rolling circle amplification. RSC Adv 5:4019–4025
Ali MM, Li F, Zhang Z, Zhang K, Kang D-K, Ankrum JA, Le XC, Zhao W (2014) Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem Soc Rev 43:3324–3341
Qu X, Bian F, Guo Q, Ge Q, Sun Q, Huang X (2018) Ligation-rolling circle amplification on quantum dot-encoded microbeads for detection of multiplex G-quadruplex-forming sequences. Anal Chem 90:12051–12058
Golub E, Albada HB, Liao W, Biniuri Y, Willner I (2016) Nucleoapzymes: hemin/G-quadruplex DNAzyme-aptamer binding site conjugates with superior enzyme-like catalytic functions. J Am Chem Soc 138:164–172
Gao Y, Li B (2014) Exonuclease III-assisted cascade signal amplification strategy for label-free and ultrasensitive chemiluminescence detection of DNA. Anal Chem 86:8881–8887
Cheglakov Z, Weizmann Y, Basnar B, Willner I (2007) Diagnosing viruses by the rolling circle amplified synthesis of DNAzymes. Org Biomol Chem 5:223–225
Xu L, Jiang Z, Mu Y, Zhang Y, Zhan Q, Cui J, Cheng W, Ding S (2018) Colorimetric assay of rare disseminated tumor cells in real sample by aptamer-induced rolling circle amplification on cell surface. Sens Actuators B Chem 259:596–603
Tian Y, He Y, Mao C (2006) Cascade signal amplification for DNA detection. ChemBioChem 7:1862–1864
Wang F, Lu CH, Liu X, Freage L, Willner I (2014) Amplified and multiplexed detection of DNA using the dendritic rolling circle amplified synthesis of DNAzyme reporter units. Anal Chem 86:1614–1621
Wen Y, Xu Y, Mao X, Wei Y, Song H, Chen N, Huang Q, Fan C, Li D (2012) DNAzyme-based rolling-circle amplification DNA machine for ultrasensitive analysis of microRNA in Drosophila larva. Anal Chem 84:7664–7669
Zhang P, Wu X, Yuan R, Chai Y (2015) An "off−on" electrochemiluminescent biosensor based on DNAzyme-assisted target recycling and rolling circle amplifications for ultrasensitive detection of microRNA. Anal Chem 87:3202–3207
Gomez A, Miller NS, Smolina I (2014) Visual detection of bacterial pathogens via PNA-based padlock probe assembly and isothermal amplification of DNAzymes. Anal Chem 86:11992–11998
Dong H, Wang C, Xiong Y, Lu H, Ju H, Zhang X (2013) Highly sensitive and selective chemiluminescent imaging for DNA detection by ligation-mediated rolling circle amplified synthesis of DNAzyme. Biosens Bioelectron 41:348–353
Rajendran A, Endo M, Hidaka K, Sugiyama H (2014) Direct and single-molecule visualization of the solution-state structures of G-hairpin and G-triplex intermediates. Angew Chem Int Ed 53:4107–4112
Wang S, Fu B, Peng S, Zhang X, Tian T, Zhou X (2013) The G-triplex DNA could function as a new variety of DNA peroxidase. Chem Commun 49:7920–7922
Wang S, Fu B, Wang J, Long Y, Zhang X, Peng S, Guo P, Tian T, Zhou X (2014) Novel amplex red oxidases based on noncanonical DNA structures: property studies and applications in microRNA detection. Anal Chem 86:2925–2930
Ma DL, Lu L, Lin S, He B, Leung CH (2015) A G-triplex luminescent switch-on probe for the detection of mung bean nuclease activity. J Mater Chem B 3:348–352
Zhou H, Wu ZF, Han QJ, Zhong HM, Peng JB, Li X, Fan XL (2018) Stable and label-free fluorescent probe based on G-triplex DNA and thioflavin T. Anal Chem 90:3220–3226
Li R, Liu Q, Jin Y, Li B (2019) G-triplex/hemin DNAzyme: an ideal signal generator for isothermal exponential amplification reaction-based biosensing platform. Anal Chim Acta 1079:139–145
Ma L, Han X, Xia L, Kong RM, Qu F (2018) G-triplex based molecular beacon for label-free fluorescence "turn-on" detection of bleomycin. Analyst 143:5474–5480
Kong D-M, Cai L-L, Guo J-H, Wu J, Shen H-X (2009) Characterization of the G-quadruplex structure of a catalytic DNA with peroxidase activity. Biopolymers 91:331–339
Xiao CD, Shibata T, Yamamoto Y, Xu Y (2018) An intramolecular antiparallel G-quadruplex form by human telomere RNA. Chem Comm 54:3994–3946
Tang L, Li J (2017) Plasmon-based colorimetric nanosensors for ultrasensitive molecular diagnostics. ACS Sens 2:857–875
Gao ZQ, Deng HM, Shen W, Ren YQ (2013) A label-free biosensor for electrochemical detection of femtomolar microRNAs. Anal Chem 85:1624–1630
Zhang Q, Chen F, Xu F, Zhao YX, Fan CH (2014) Target-triggered three-way junction structure and polymerase/nicking enzyme synergetic isothermal quadratic DNA machine for highly specific, one-step, and rapid microRNA detection at attomolar level. Anal Chem 86:8098–8105
Lin Y, Zhao J, Hu X, Wang L, Liang L, Chen W (2016) Transcription factor CCAAT/enhancer binding protein alpha up-regulates microRNA let-7a-1 in lung cancer cells by direct binding. Cancer Cell Int 16:17
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 21775099 and 21974082).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 4.28 mb)
Rights and permissions
About this article
Cite this article
Li, R., Liu, Q., Jin, Y. et al. Sensitive colorimetric determination of microRNA let-7a through rolling circle amplification and a peroxidase-mimicking system composed of trimeric G-triplex and hemin DNAzyme. Microchim Acta 187, 139 (2020). https://doi.org/10.1007/s00604-019-4093-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-4093-2