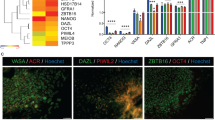
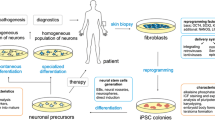
Abstract
Purpose
The expansion of CAG (glutamine; Q) trinucleotide repeats (TNRs) predominantly occurs through male lineage in Huntington’s disease (HD). As a result, offspring will have larger CAG repeats compared to their fathers, which causes an earlier onset of the disease called genetic anticipation. This study aims to develop a novel in vitro model to replicate CAG repeat instability in early spermatogenesis and demonstrate the biological process of genetic anticipation by using the HD stem cell model for the first time.
Methods
HD rhesus monkey embryonic stem cells (rESCs) were cultured in vitro for an extended period. Male rESCs were used to derive spermatogenic cells in vitro with a 10-day differentiation. The assessment of CAG repeat instability was performed by GeneScan and curve fit analysis.
Results
Spermatogenic cells derived from rESCs exhibit progressive expansion of CAG repeats with high daily expansion rates compared to the extended culture of rESCs. The expansion of CAG repeats is cell type–specific and size-dependent.
Conclusions
Here, we report a novel stem cell model that replicates genome instability and CAG repeat expansion in in vitro derived HD monkey spermatogenic cells. The in vitro spermatogenic cell model opens a new opportunity for studying TNR instability and the underlying mechanism of genetic anticipation, not only in HD but also in other TNR diseases.






Similar content being viewed by others
Data availability
N/A
Code availability
N/A
References
Bates GP, Dorsey R, Gusella JF, Hayden MR, Kay C, Leavitt BR, et al. Huntington disease. Nat Rev Dis Primers. 2015;1:15005.
Ghosh R, Tabrizi SJ. Clinical features of Huntington’s disease. Adv Exp Med Biol. 2018;1049:1–28.
Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, Warner JH, et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol. 2014;10(4):204–16.
Aronin N, DiFiglia M. Huntingtin-lowering strategies in Huntington’s disease: antisense oligonucleotides, small RNAs, and gene editing. Mov Disord. 2014;29(11):1455–61.
Didiot MC, Hall LM, Coles AH, Haraszti RA, Godinho BM, Chase K, et al. Exosome-mediated delivery of hydrophobically modified siRNA for Huntingtin mRNA silencing. Mol Ther. 2016;24(10):1836–47.
Gagnon KT, Pendergraff HM, Deleavey GF, Swayze EE, Potier P, Randolph J, et al. Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49(47):10166–78.
Johnson E, Chase K, McGowan S, Mondo E, Pfister E, Mick E, et al. Safety of striatal infusion of siRNA in a transgenic Huntington’s disease mouse model. J Huntingtons Dis. 2015;4(3):219–29.
Skotte NH, Southwell AL, Ostergaard ME, Carroll JB, Warby SC, Doty CN, et al. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all Huntington disease patients. PLoS One. 2014;9(9):e107434.
Yang S, Chang R, Yang H, Zhao T, Hong Y, Kong HE, et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J Clin Invest. 2017;127(7):2719–24.
McColgan P, Tabrizi SJ. Huntington’s disease: a clinical review. Eur J Neurol. 2018;25(1):24–34.
Rawlins MD, Wexler NS, Wexler AR, Tabrizi SJ, Douglas I, Evans SJ, et al. The prevalence of Huntington’s disease. Neuroepidemiology. 2016;46(2):144–53.
Cannella M, Maglione V, Martino T, Ragona G, Frati L, Li GM, et al. DNA instability in replicating Huntington’s disease lymphoblasts. BMC Med Genet. 2009;10:11.
Clever F, Cho IK, Yang J, Chan AWS. Progressive polyglutamine repeat expansion in peripheral blood cells and sperm of transgenic Huntington’s disease monkeys. J Huntingtons Dis. 2019;8(4):443–8.
Hung CL, Maiuri T, Bowie LE, Gotesman R, Son S, Falcone M, et al. A patient-derived cellular model for Huntington’s disease reveals phenotypes at clinically relevant CAG lengths. Mol Biol Cell. 2018;29(23):2809–20.
Jacquet L, Neueder A, Foldes G, Karagiannis P, Hobbs C, Jolinon N, et al. Three Huntington’s disease specific mutation-carrying human embryonic stem cell lines have stable number of CAG repeats upon in vitro differentiation into cardiomyocytes. PLoS One. 2015;10(5):e0126860.
Manley K, Pugh J, Messer A. Instability of the CAG repeat in immortalized fibroblast cell cultures from Huntington’s disease transgenic mice. Brain Res. 1999;835(1):74–9.
McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11(11):786–99.
Mollica PA, Reid JA, Ogle RC, Sachs PC, Bruno RD. DNA Methylation leads to DNA repair gene down-regulation and trinucleotide repeat expansion in patient-derived Huntington disease cells. Am J Pathol. 2016;186(7):1967–76.
Mollica PA, Zamponi M, Reid JA, Sharma DK, White AE, Ogle RC, et al. Epigenetic alterations mediate iPSC-induced normalization of DNA repair gene expression and TNR stability in Huntington's disease cells. J Cell Sci. 2018;131(13).
Simard O, Gregoire MC, Arguin M, Brazeau MA, Leduc F, Marois I, et al. Instability of trinucleotidic repeats during chromatin remodeling in spermatids. Hum Mutat. 2014;35(11):1280–4.
Usdin K, House NC, Freudenreich CH. Repeat instability during DNA repair: insights from model systems. Crit Rev Biochem Mol Biol. 2015;50(2):142–67.
Yoon SR, Dubeau L, de Young M, Wexler NS, Arnheim N. Huntington disease expansion mutations in humans can occur before meiosis is completed. Proc Natl Acad Sci U S A. 2003;100(15):8834–8.
Heitz D, Devys D, Imbert G, Kretz C, Mandel JL. Inheritance of the fragile X syndrome: size of the fragile X premutation is a major determinant of the transition to full mutation. J Med Genet. 1992;29(11):794–801.
Sherman SL, Jacobs PA, Morton NE, Froster-Iskenius U, Howard-Peebles PN, Nielsen KB, et al. Further segregation analysis of the fragile X syndrome with special reference to transmitting males. Hum Genet. 1985;69(4):289–99.
Jones L, Houlden H, Tabrizi SJ. DNA repair in the trinucleotide repeat disorders. Lancet Neurol. 2017;16(1):88–96.
Murmann AE, Yu J, Opal P, Peter ME. Trinucleotide repeat expansion diseases, RNAi, and cancer. Trends Cancer. 2018;4(10):684–700.
Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621.
Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27(4):407–11.
Kraus-Perrotta C, Lagalwar S. Expansion, mosaicism and interruption: mechanisms of the CAG repeat mutation in spinocerebellar ataxia type 1. Cerebellum Ataxias. 2016;3:20.
Massey T, McAllister B, Jones L. Methods for assessing DNA repair and repeat expansion in Huntington’s disease. Methods Mol Biol. 1780;2018:483–95.
Mollersen L, Rowe AD, Larsen E, Rognes T, Klungland A. Continuous and periodic expansion of CAG repeats in Huntington’s disease R6/1 mice. PLoS Genet. 2010;6(12):e1001242.
Snell RG, MacMillan JC, Cheadle JP, Fenton I, Lazarou LP, Davies P, et al. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet. 1993;4(4):393–7.
Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, Frontali M, et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet. 1993;4(4):387–92.
Kremer B, Almqvist E, Theilmann J, Spence N, Telenius H, Goldberg YP, et al. Sex-dependent mechanisms for expansions and contractions of the CAG repeat on affected Huntington disease chromosomes. Am J Hum Genet. 1995;57(2):343–50.
MacDonald ME, Barnes G, Srinidhi J, Duyao MP, Ambrose CM, Myers RH, et al. Gametic but not somatic instability of CAG repeat length in Huntington’s disease. J Med Genet. 1993;30(12):982–6.
Myers RH, MacDonald ME, Koroshetz WJ, Duyao MP, Ambrose CM, Taylor SA, et al. De novo expansion of a (CAG)n repeat in sporadic Huntington’s disease. Nat Genet. 1993;5(2):168–73.
Ranen NG, Stine OC, Abbott MH, Sherr M, Codori AM, Franz ML, et al. Anticipation and instability of IT-15 (CAG)n repeats in parent-offspring pairs with Huntington disease. Am J Hum Genet. 1995;57(3):593–602.
Telenius H, Almqvist E, Kremer B, Spence N, Squitieri F, Nichol K, et al. Somatic mosaicism in sperm is associated with intergenerational (CAG)n changes in Huntington disease. Hum Mol Genet. 1995;4(2):189–95.
Ehmcke J, Wistuba J, Schlatt S. Spermatogonial stem cells: questions, models and perspectives. Hum Reprod Update. 2006;12(3):275–82.
Fayomi AP, Orwig KE. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. 2018;29:207–14.
McCarrey JR. EPIGENETICS. The epigenome--a family affair. Science. 2015;350(6261):634–5.
McMurray CT, Kortun IV. Repair in haploid male germ cells occurs late in differentiation as chromatin is condensing. Chromosoma. 2003;111(8):505–8.
Neto JL, Lee JM, Afridi A, Gillis T, Guide JR, Dempsey S, et al. Genetic contributors to intergenerational CAG repeat instability in Huntington’s disease knock-in mice. Genetics. 2017;205(2):503–16.
Aziz NA, van Belzen MJ, Coops ID, Belfroid RD, Roos RA. Parent-of-origin differences of mutant HTT CAG repeat instability in Huntington’s disease. Eur J Med Genet. 2011;54(4):e413–8.
Norremolle A, Sorensen SA, Fenger K, Hasholt L. Correlation between magnitude of CAG repeat length alterations and length of the paternal repeat in paternally inherited Huntington's disease. Clin Genet. 1995;47(3):113–7.
Hermann BP, Cheng K, Singh A, Roa-De La Cruz L, Mutoji KN, Chen IC, et al. The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 2018;25(6):1650–67.e8.
Luetjens CM, Weinbauer GF, Wistuba J. Primate spermatogenesis: new insights into comparative testicular organisation, spermatogenic efficiency and endocrine control. Biol Rev Camb Philos Soc. 2005;80(3):475–88.
Putkhao K, Kocerha J, Cho IK, Yang J, Parnpai R, Chan AW. Pathogenic cellular phenotypes are germline transmissible in a transgenic primate model of Huntington’s disease. Stem Cells Dev. 2013;22(8):1198–205.
Yang SH, Cheng PH, Banta H, Piotrowska-Nitsche K, Yang JJ, Cheng EC, et al. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature. 2008;453(7197):921–4.
Easley CA, Phillips BT, McGuire MM, Barringer JM, Valli H, Hermann BP, et al. Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep. 2012;2(3):440–6.
Gazy I, Hayward B, Potapova S, Zhao X, Usdin K. Double-strand break repair plays a role in repeat instability in a fragile X mouse model. DNA Repair. 2019;74:63–9.
Zhao X, Zhang Y, Wilkins K, Edelmann W, Usdin K. MutLγ promotes repeat expansion in a Fragile X mouse model while EXO1 is protective. PLoS Genet. 2018;14(10):e1007719.
Chan AW, Xu Y, Jiang J, Rahim T, Zhao D, Kocerha J, et al. A two years longitudinal study of a transgenic Huntington disease monkey. BMC Neurosci. 2014;15:36.
Mollersen L, Rowe AD, Illuzzi JL, Hildrestrand GA, Gerhold KJ, Tveteras L, et al. Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice. Hum Mol Genet. 2012;21(22):4939–47.
Steves AN, Bradner JM, Fowler KL, Clarkson-Townsend D, Gill BJ, Turry AC, et al. Ubiquitous flame-retardant toxicants impair spermatogenesis in a human stem cell model. iScience. 2018;3:161–76.
Steves AN, Turry A, Gill B, Clarkson-Townsend D, Bradner JM, Bachli I, et al. Per- and polyfluoroalkyl substances impact human spermatogenesis in a stem-cell-derived model. Syst Biol Reprod Med. 2018;64(4):225–39.
Chan AW, Jiang J, Chen Y, Li C, Prucha MS, Hu Y, et al. Progressive cognitive deficit, motor impairment and striatal pathology in a transgenic Huntington disease monkey model from infancy to adulthood. PLoS One. 2015;10(5):e0122335.
Kocerha J, Liu Y, Willoughby D, Chidamparam K, Benito J, Nelson K, et al. Longitudinal transcriptomic dysregulation in the peripheral blood of transgenic Huntington's disease monkeys. BMC Neurosci. 2013;14:88.
Lallani SB, Villalba RM, Chen Y, Smith Y, Chan AWS. Striatal interneurons in transgenic nonhuman primate model of Huntington’s disease. Sci Rep. 2019;9(1):3528.
Meng Y, Jiang J, Bachevalier J, Zhang X, Chan AW. Developmental whole brain white matter alterations in transgenic Huntington’s disease monkey. Sci Rep. 2017;7(1):379.
Raper J, Bosinger S, Johnson Z, Tharp G, Moran SP, Chan AW. Increased irritability, anxiety, and immune reactivity in transgenic Huntington’s disease monkeys. Brain Behav Immun. 2016;58:181–90.
MacRae SL, Croken MM, Calder RB, Aliper A, Milholland B, White RR, et al. DNA repair in species with extreme lifespan differences. Aging (Albany NY). 2015;7(12):1171–84.
Mouse Genome Sequencing C, Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–62.
Lai Y, Budworth H, Beaver JM, Chan NL, Zhang Z, McMurray CT, et al. Crosstalk between MSH2-MSH3 and polbeta promotes trinucleotide repeat expansion during base excision repair. Nat Commun. 2016;7:12465.
Massey TH, Jones L. The central role of DNA damage and repair in CAG repeat diseases. Dis Model Mech. 2018;11(1).
Wheeler VC, Lebel LA, Vrbanac V, Teed A, te Riele H, MacDonald ME. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum Mol Genet. 2003;12(3):273–81.
Pinto RM, Dragileva E, Kirby A, Lloret A, Lopez E, St Claire J, et al. Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington's disease mice: genome-wide and candidate approaches. PLoS Genet. 2013;9(10):e1003930.
Ooi J, Langley SR, Xu X, Utami KH, Sim B, Huang Y, et al. Unbiased profiling of isogenic huntington disease hPSC-derived CNS and peripheral cells reveals strong cell-type specificity of CAG length effects. Cell Rep. 2019;26(9):2494–508.e7.
Cardozo-Pelaez F, Song S, Parthasarathy A, Hazzi C, Naidu K, Sanchez-Ramos J. Oxidative DNA damage in the aging mouse brain. Mov Disord. 1999;14(6):972–80.
Goold R, Flower M, Moss DH, Medway C, Wood-Kaczmar A, Andre R, et al. FAN1 modifies Huntington’s disease progression by stabilizing the expanded HTT CAG repeat. Hum Mol Genet. 2019;28(4):650–61.
Jonson I, Ougland R, Klungland A, Larsen E. Oxidative stress causes DNA triplet expansion in Huntington’s disease mouse embryonic stem cells. Stem Cell Res. 2013;11(3):1264–71.
Maiuri T, Mocle AJ, Hung CL, Xia J, van Roon-Mom WM, Truant R. Huntingtin is a scaffolding protein in the ATM oxidative DNA damage response complex. Hum Mol Genet. 2017;26(2):395–406.
Ferlazzo ML, Sonzogni L, Granzotto A, Bodgi L, Lartin O, Devic C, et al. Mutations of the Huntington’s disease protein impact on the ATM-dependent signaling and repair pathways of the radiation-induced DNA double-strand breaks: corrective effect of statins and bisphosphonates. Mol Neurobiol. 2014;49(3):1200–11.
Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17(6):679–87.
Budworth H, McMurray CT. A brief history of triplet repeat diseases. Methods Mol Biol. 2013;1010:3–17.
Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460(7254):473–8.
Kovtun IV, Goellner G, McMurray CT. Structural features of trinucleotide repeats associated with DNA expansion. Biochem Cell Biol. 2001;79(3):325–36.
Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 2015;350(6261):aab2006.
Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36(6):647–52.
Hermann BP, Sukhwani M, Simorangkir DR, Chu T, Plant TM, Orwig KE. Molecular dissection of the male germ cell lineage identifies putative spermatogonial stem cells in rhesus macaques. Hum Reprod. 2009;24(7):1704–16.
Kovtun IV, Therneau TM, McMurray CT. Gender of the embryo contributes to CAG instability in transgenic mice containing a Huntington’s disease gene. Hum Mol Genet. 2000;9(18):2767–75.
Telenius H, Kremer B, Goldberg YP, Theilmann J, Andrew SE, Zeisler J, et al. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet. 1994;6(4):409–14.
Wheeler VC, Persichetti F, McNeil SM, Mysore JS, Mysore SS, MacDonald ME, et al. Factors associated with HD CAG repeat instability in Huntington disease. J Med Genet. 2007;44(11):695–701.
Beaver JM, Lai Y, Xu M, Casin AH, Laverde EE, Liu Y. AP endonuclease 1 prevents trinucleotide repeat expansion via a novel mechanism during base excision repair. Nucleic Acids Res. 2015;43(12):5948–60.
Crespan E, Hubscher U, Maga G. Expansion of CAG triplet repeats by human DNA polymerases lambda and beta in vitro, is regulated by flap endonuclease 1 and DNA ligase 1. DNA Repair (Amst). 2015;29:101–11.
Mason AG, Tome S, Simard JP, Libby RT, Bammler TK, Beyer RP, et al. Expression levels of DNA replication and repair genes predict regional somatic repeat instability in the brain but are not altered by polyglutamine disease protein expression or age. Hum Mol Genet. 2014;23(6):1606–18.
Liu Y, Zhang Y, Yin J, Gao Y, Li Y, Bai D, et al. Distinct H3K9me3 and DNA methylation modifications during mouse spermatogenesis. J Biol Chem. 2019;294(49):18714–25.
Lombo M, Fernandez-Diez C, Gonzalez-Rojo S, Herraez MP. Genetic and epigenetic alterations induced by bisphenol A exposure during different periods of spermatogenesis: from spermatozoa to the progeny. Sci Rep. 2019;9(1):18029.
McSwiggin HM, O'Doherty AM. Epigenetic reprogramming during spermatogenesis and male factor infertility. Reproduction. 2018;156(2):R9–R21.
Acknowledgements
All materials and experiments were performed at the Yerkes National Primate Research Center (YNPRC). We would like to thank YNPRC staff, Jinjing Yang, Siran Tian, Dr. Kanchana Punyawai, and other Dr. Chan’s Lab members.
Funding
YNPRC is supported by the Office of Research and Infrastructure Program (ORIP)/OD P51OD11132. The Transgenic Huntington’s Disease Monkey Resource “THDMR” and this study were supported in part by grants awarded by the ORIP/NIH (OD010930) and NINDS/NIH (NS101701) to AWSC. This study was also supported in part by OD020182 and Georgia Partners in Regenerative Medicine Seed Grant to AWSC and CAE. We also received the support by the Emory University Research Council and Arthur and Sarah Merrill Foundation to AWSC and IKC. Sujittra Khampang and Rangsun Parnpai are supported by SUT-PhD scholarship, Suranaree University of Technology, Nakhorachasrima, Thailand.
Author information
Authors and Affiliations
Contributions
Anthony W. S. Chan, In Ki Cho, and Sujittra Khampang conceptualized the study, designed experimentations, and wrote the manuscript. Sujittra Khampang and In Ki Cho performed experiments, data collection, and data analysis. Anthony W. S. Chan and Charles A. Easley IV supervised the spermatogenic cell differentiation, characterization, and experimental procedures. Wiriya Mahikul supervised the data analysis and statistical interpretation. Rangsun Parnpai supervised and reviewed the manuscript. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Disclaimer
This work was prepared while Anthony W.S. Chan was employed at Yerkes National Primate Research Center and Emory University. The opinions expressed in this article are the author's own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States government.
Ethics approval
N/A
Consent to participate
N/A
Consent for publication
N/A
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 2468 kb)
Rights and permissions
About this article
Cite this article
Khampang, S., Parnpai, R., Mahikul, W. et al. CAG repeat instability in embryonic stem cells and derivative spermatogenic cells of transgenic Huntington’s disease monkey. J Assist Reprod Genet 38, 1215–1229 (2021). https://doi.org/10.1007/s10815-021-02106-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-021-02106-3