Abstract
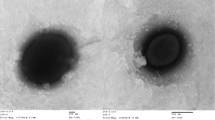
Genome analysis of Halomonas shambharensis, a novel species, was performed to understand the osmoprotectant strategies used by the strain to overcome the salinity stress and to explore the prospective industrial uses. It will also help to better understand the ecological roles of Halomonas species in hypersaline habitats. Ultrastructure of the cell was determined by using transmission electron microscopy. Standard microbiological methods were used to find out growth parameters and heterotrophic mode of nutrition. For Genome analysis, complete bacterial genome sequencing was performed using the Oxford Nanopore MinION DNA Sequencer. Assembly, annotation and finishing of the obtained sequence were done by using a Prokaryotic Genome Annotation Pipeline (PGAP) (SPAdes v. 3.10.1). Predicted Coading sequences (CDSs) obtained through the PGAP were used for functional annotation using Clusters of Orthologous Groups and Kyoto Encyclopedia of Genes and Genomes (KEGG) platforms. The H. shambharensis was found to be a Gram-stain-negative, rod-shaped bacterium, motile with a peritrichous flagella. The H. shambharensis bacterium can grow in a wide range of temperature (from 25 to 65 °C), pH (pH 4 to pH 12.0) and salt concentration (5.0% NaCl to 30.0% NaCl). After annotation and assembly, the total genome size obtained was 1,533,947 bp, which revealed 146 subsystems, 3847 coding sequences, and 19RNAs with G+C content of 63.6%. Gene annotation identified the genes related to various metabolic pathways, including carbohydrate metabolism, fatty acid metabolism and stress tolerance. The genomic dataset of H. shambharensis will be useful for analysis of protein-coding gene families and how these coding genes are significant for the survival and metabolism among the different species of Halomonas. The complete genome sequence presented here will help to unravel the biotechnological potential of H. shambharensis for production of the high-value products such as betaine, or as a source of gene-mining for individual enzymes.




Similar content being viewed by others
Data availability
The genome sequence and associated data for Halomonas sp. SBS 10 was deposited under GenBank accession number RXHI00000000, BioProject accession number PRJNA479678, SRA accession number SRP224914, and BioSample accession number SAMN09601649.
References
Arahal DR, Ventosa A (2006) The family Halomonadaceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K.-H., Stackebrandt E (eds) The prokaryotes, Springer, New York, pp 811–835.
de la Haba RR, Sánchez-Porro C, Ventosa A (2011) Taxonomy, phylogeny, and biotechnological interest of the family Halomonadaceae. In: Ventosa A, Oren A, Ma Y (eds) Halophiles and hypersaline environments. Springer, Heidelberg, pp 27–64
Lentzen G, Schwarz T (2006) Extremolytes: natural compounds from extremophiles for versatile applications. Appl Microbiol Biotechnol 72:623–634
Jadhav K, Kushwah B, Jadhav I (2018) Insight into compatible solutes from halophiles: exploring significant applications in biotechnology. In: Singh J, Sharma D, Kumar G, Sharma N (eds) Microbial bioprospecting for sustainable development. Springer-Nature, Singapore, pp 291–307. https://doi.org/10.1007/978-981-13-0053-0_16
Satpute SK, Banat IM, Dhakephalkar PK, Banpurkar AG, Chopade BA (2010) Biosurfactants, bioemulsifiers and exopolysaccharides from marine microorganisms. Biotechnol Adv 28:436–450
Sharma A, Paul D, Dhotre D, Jani K, Pandey A, Shouche Y (2017) Deep sequencing analysis of bacterial community structure of Soldhar hot spring, India. Microbiology 86:126–132
Kushwaha B, Sharma GP, Sharma A, Shankar P, Geethadevi A, Sharma N, Sharma MK, Jadhav I, Parashar D, Jadhav K (2020a) Whole-genome shotgun sequence of Halomonas sp. strain SBS 10, isolated from a hypersaline lake in India. Microbiol Resour Announce 9:e01270. https://doi.org/10.1128/MRA.01270-19
Kushwaha B, Jadhav I, Jadhav K (2020b) Halomonas sambharensis sp. nov., a moderately halophilic bacterium isolated from the saltern crystallizer ponds of the Sambhar Salt Lake in India. Curr Microbiol 77:1125. https://doi.org/10.1007/s00284-020-01892-w
Gao XY, Zhi XY, Li HW, Zhou Y, Lapidus A, Han J, Haynes M, Lobos E, Huntemann M, Pati A, Ivanova NN, Mavromatis K, Tindall BJ, Markowitz V, Woyke T, Klenk HP, Kyrpides NC, Li WJ (2015) Draft genome sequence of Halomonas lutea strain YIM 91125(T) (DSM 23508(T)) isolated from the alkaline Lake Ebinur in Northwest China. Stand Genomic Sci 10:1. https://doi.org/10.1186/1944-3277-10-1
Jadhav K, Jadhav I (2017) Sulfur oxidation by Achromobacter xylosoxidans strain wsp05 reveals ecological widening over which thiotrophs are distributed. World J Microbiol Biotechnol 30:192. https://doi.org/10.1007/s11274-017-2359-6
Tatiana T, Anne E, Alexis B, Stéphane B (2019) Complete genome sequence of the halophilic PHA-producing bacterium Halomonas sp. SF2003: insights into its biotechnological potential. World J Microbiol Biotechnol 35:50. https://doi.org/10.1007/s11274-019-2627-8
Williamson A, De Santi C, Altermark B, Karlsen C, Hjerde E (2016) Complete genome sequence of Halomonas sp. R5-57. Stand Genomic Sci 11(1):62. https://doi.org/10.1186/s40793-016-0192-4
Ventosa A, Quesada E, Rodrı´guez-Valera F, Ruiz-Berraquero F, Ramos-Cormenzana A (1982) Numerical taxonomy of moderately halophilic Gram-negative rods. J Gen Microbiol 128:959–1968
Bouchotroch S, Quesada E, Ad M, Llamas I, Bejar V (2001) Halomonasmaura sp. nov., a novel moderately halophilic, exopolysaccharide-producing bacterium. Int J Syst Evol Microbiol 51:1625–1632
Kushwah B, Jadhav I, Verma HN, Geethadevi A, Parashar D, Jadhav K (2019) Betaine accumulation suppresses the de-novo synthesis of ectoine at a low osmotic concentration in Halomonas sp SBS 10, a bacterium with broad salinity tolerance. Mol Biol Rep 46(5):4779–4786. https://doi.org/10.1007/s11033-019-04924-2
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Mukherjee S, Stamatis S, Bertsch J, Ovchinnikova G, Katta HY, Mojica A, Chen AMI, Kyrpides CN, Reddy TBK (2018) Genomes OnLine database (GOLD) v.7: updates and new features. Nucleic Acids Res 47:D649. https://doi.org/10.1093/nar/gky977
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, De Vos P, DePamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glöckner FO, Goldstein P, Guralnick R, Haft D, Hancock D et al (2008) The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 26:541–547
Haft DH, DiCuccio M, Badretdin A, Brover V, Chetvernin V, O’Neill K, Li W, Chitsaz F, Derbyshire MK, Gonzales NR, Gwadz M, Lu F, Marchler GH, Song JS, Thanki N, Yamashita RA, Zheng C, Thibaud-Nissen F, Geer LY, Marchler-Bauer A, Pruitt KD (2018) RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res 46(1):851–860. https://doi.org/10.1093/nar/gkx1068
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44(14):6614–6624. https://doi.org/10.1093/nar/gkw569
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. https://doi.org/10.1186/1471-2164-9-75
Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F (2015) RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. https://doi.org/10.1038/srep08365
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206. https://doi.org/10.1093/nar/gkt1226
Vallenet D, Calteau A, Cruveiller S, Gachet M, Lajus A, Josso A, Mercier J, Renaux A, Rollin A, Rouy Z, Roche D, Scarpelli C, Médigue C (2017) MicroScope: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res 45(1):517–528. https://doi.org/10.1093/nar/gkw1101
Woese CR, Kandler O, Wheelis ML (1990) Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eucarya. Proc Natl Acad Sci 87:4576–4579
Garrity GM, Bell JA, Lilburn T (2005) Oceanospirillales ord. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, Boone DR, De Vos P (eds) Bergey’s Manual® of systematic bacteriology: volume two the Proteobacteria part B the Gammaproteobacteria. Springer, Boston, pp 270–323
Franzmann PD, Wehmeyer U, Stackebrandt E (1988) Halomonadaceae fam. nov., a new family of the class Proteobacteria to accommodate the genera Halomonas and Deleya. Syst Appl Microbiol 11:16–19
Vreeland RH, Litchfield CD, Martin EL, Elliot E (1980) Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Int J Syst Evol Microbiol 30:485–495
Mata JA, Martinez-Canovas J, Quesada E, Bejar V (2002) A detailed phenotypic characterisation of the type strains of Halomonas species. Syst Appl Microbiol 25:360–375
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28(1):33–36. https://doi.org/10.1093/nar/28.1.33
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M (2019) New approach for understanding genome variations in KEGG. Nucleic Acids Res 47:D590–D595
Galinski EA (1995) Osmoadaptation in bacteria. Adv Microb Physiol 37:272–328
Bohin JP (2000) Osmoregulated periplasmic glucans in Proteobacteria. FEMS Lett 186:11–19
Daily OP, Debell RM, Joseph SW (1978) Superoxide dismutase and catalase levels in halophilic vibrios. J Bacteriol 134(2):375–380
Mishra S, Imlay J (2012) Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys 525(2):145–160. https://doi.org/10.1016/j.abb.2012.04.014
Meyer JL, Dillard BA, Rodgers JM, Ritchie KB, Paul VJ, Teplitski M (2015) Draft genome sequence of Halomonas meridiana R1t3 isolated from the surface microbiota of the Caribbean Elkhorn coral Acropora palmata. Stand Genomic Sci 10:75
Kottemann M, Kish A, Iloanusi C, Bjork S, DiRuggiero J (2005) Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles 9:219–227. https://doi.org/10.1007/s00792-005-0437-4
Oren A (2015) Pyruvate: a key nutrient in hypersaline environments? Microorganisms 3(3):407–416
Garrity GM, Bell JA, Lilburn T. Class I (2005) Alphaproteobacteria class. Nov. In: Brenner DJ, Krieg NR, Staley JT (eds) Bergey’s manual of systematic bacteriology: volume two, the Proteobacteria part C the alpha-, beta-, delta-, and epsilonproteobacteria, Springer , Boston , US, pp 1–574.
León MJ, Hoffmann T, Sánchez-Porro C, Heider J, Ventosa A, Bremer E (2018) Compatible solute synthesis and import by the moderate halophile Spiribacter salinus: physiology and genomics. Front Microbiol 9:108. https://doi.org/10.3389/fmicb.2018.00108
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jadhav, K., Kushwaha, B., Jadhav, I. et al. Genomic analysis of a novel species Halomonas shambharensis isolated from hypersaline lake in Northwest India. Mol Biol Rep 48, 1045–1053 (2021). https://doi.org/10.1007/s11033-020-06131-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-06131-w