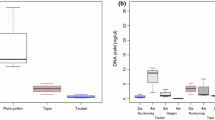
Abstract
Pollen metabarcoding is emerging as a powerful tool for ecological research and offers unprecedented scale in citizen science projects for environmental monitoring via honey bees. Biases in metabarcoding can be introduced at any stage of sample processing and preservation is at the forefront of the pipeline. While in metabarcoding studies pollen has been preserved at − 20 °C (FRZ), this is not the best method for citizen scientists. Herein, we compared this method with ethanol (EtOH), silica gel (SG) and room temperature (RT) for preservation of pollen collected from hives in Austria and Denmark. After ~ 4 months of storage, DNAs were extracted with a food kit, and their quality and concentration measured. Most DNA extracts exhibited 260/280 absorbance ratios close to the optimal 1.8, with RT samples from Austria performing slightly worse than FRZ and SG samples (P < 0.027). Statistical differences were also detected for DNA concentration, with EtOH samples producing lower yields than RT and FRZ samples in both countries and SG in Austria (P < 0.042). Yet, qualitative and quantitative assessments of floral composition obtained using high-throughput sequencing with the ITS2 barcode gave non-significant effects of preservation methods on richness, relative abundance and Shannon diversity, in both countries. While freezing and ethanol are commonly employed for archiving tissue for molecular applications, desiccation is cheaper and easier to use regarding both storage and transportation. Since SG is less dependent on ambient humidity and less prone to contamination than RT, we recommend SG for preserving pollen for metabarcoding. SG is straightforward for laymen to use and hence robust for widespread application in citizen science studies.









Similar content being viewed by others
Data availability
All source data analysed is available in the supplemental material.
References
Alaux, C., Ducloz, F., Crauser, D., & Le Conte, Y. (2010). Diet effects on honeybee immunocompetence. Biology Letters, 6(4), 562–565. https://doi.org/10.1098/rsbl.2009.0986
Alsos, I. G., Lavergne, S., Merkel, M. K., Boleda, M., Lammers, Y., Alberti, A., Pouchon, C., Denoeud, F., Pitelkova, I., Pușcaș, M., Roquet, C., Hurdu, B.-I., Thuiller, W., Zimmermann, N. E., Hollingsworth, P. M., & Coissac, E. (2020). The treasure vault can be opened: large-scale genome skimming works well using herbarium and silica gel dried material. Plants, 9(4). https://doi.org/10.3390/plants9040432
Avni, D., Hendriksma, H. P., Dag, A., Uni, Z., & Shafir, S. (2014). Nutritional aspects of honey bee-collected pollen and constraints on colony development in the eastern Mediterranean. Journal of Insect Physiology, 69, 65–73. https://doi.org/10.1016/j.jinsphys.2014.07.001
Banchi, E., Ametrano, C. G., Greco, S., Stanković, D., Muggia, L., & Pallavicini, A. (2020). PLANiTS: a curated sequence reference dataset for plant ITS DNA metabarcoding. Database, 2020. https://doi.org/10.1093/database/baz155
Beekman, M., & Ratnieks, F. L. W. (2000). Long-range foraging by the honey-bee, Apis mellifera L. Functional Ecology, 14, 490–496. https://doi.org/10.1046/j.1365-2435.2000.00443.x.
Bell, K. L., Burgess, K. S., Botsch, J. C., Dobbs, E. K., Read, T. D., & Brosi, B. J. (2018). Quantitative and qualitative assessment of pollen DNA metabarcoding using constructed species mixtures. Molecular Ecology, 28(2), 431–455. https://doi.org/10.1111/mec.14840
Bell, K. L., De Vere, N., Keller, A., Richardson, R. T., Gous, A., Burgess, K. S., & Brosi, B. J. (2016). Pollen DNA barcoding: Current applications and future prospects. Genome, 59, 629–640. https://doi.org/10.1139/gen-2015-0200
Bell, K. L., Fowler, J., Burgess, K. S., Dobbs, E. K., Gruenewald, D., Lawley, B., Morozumi, C., & Brosi, B. J. (2017). Applying pollen DNA metabarcoding to the study of plant–pollinator interactions. Applications in Plant Sciences, 5, 1600124. https://doi.org/10.3732/apps.1600124
Beuchat, L. R. (1983). Influence of water activity on growth, metabolic activities and survival of yeasts and molds. Journal of Food Protection, 46(2), 135–141. https://doi.org/10.4315/0362-028X-46.2.135
Bilisik, A., Cakmak, I., Bicakci, A., & Malyer, H. (2008). Seasonal variation of collected pollen loads of honeybees (Apis mellifera L. anatoliaca). Grana, 47(1), 70–77. https://doi.org/10.1080/00173130801923976
Bressan, E. A., Rossi, M. L., Gerald, L. T. S., & Figueira, A. (2014). Extraction of high-quality DNA from ethanol-preserved tropical plant tissues. BMC Research Notes, 7(1), 268. https://doi.org/10.1186/1756-0500-7-268
Brodschneider, R., & Crailsheim, K. (2010). Nutrition and health in honey bees. Apidologie, 41(3), 278–294. https://doi.org/10.1051/apido/2010012
Brodschneider, R., Gratzer, K., Carreck, N. L., Vejsnaes, F., & van der Steen, J. (2021). INSIGNIA: beekeepers as citizen scientists investigate the environment of their honey bees. Proceedings of Science. https://doi.org/10.22323/1.393.0019
Brodschneider, R., Gratzer, K., Kalcher-Sommersguter, E., Heigl, H., Auer, W., Moosbeckhofer, R., & Crailsheim, K. (2019). A citizen science supported study on seasonal diversity and monoflorality of pollen collected by honey bees in Austria. Scientific Reports, 9, 16633. https://doi.org/10.1038/s41598-019-53016-5
Brooks, J. P., Edwards, D. J., Harwich, M. D., Rivera, M. C., Fettweis, J. M., Serrano, M. G., Reris, R. A., Sheth, N. U., Huang, B., Girerd, P., Strauss, J. F., Jefferson, K. K., & Buck, G. A. (2015). The truth about metagenomics: Quantifying and counteracting bias in 16S rRNA studies. BMC Microbiology, 15(1), 66. https://doi.org/10.1186/s12866-015-0351-6
Canale, A., Benelli, G., Castagna, A., Sgherri, C., Poli, P., Serra, A., Mele, M., Ranieri, A., Signorini, F., Bientinesi, M., & Nicolella, C. (2016). Microwave-assisted drying for the conservation of honeybee pollen. Materials, 9, 363. https://doi.org/10.3390/ma9050363
Chase, M. W., & Hills, H. H. (1991). Silica gel: An ideal material for field preservation of leaf samples for DNA studies. Taxon, 40(2), 215–220. https://doi.org/10.2307/1222975
Chen, S., Yao, H., Han, J., Liu, C., Song, J., Shi, L., Zhu, Y., Ma, X., Gao, T., Pang, X., Luo, K., Li, Y., Li, X., Jia, X., Lin, Y., & Leon, C. (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE, 5(1), e8613. https://doi.org/10.1371/journal.pone.0008613
Cheng, T., Xu, C., Lei, L., Li, C., Zhang, Y., & Zhou, S. (2016). Barcoding the kingdom Plantae: New PCR primers for ITS regions of plants with improved universality and specificity. Molecular Ecology Resources, 16(1), 138–149. https://doi.org/10.1111/1755-0998.12438
Coffey, M. F., & Breen, J. (1997). Seasonal variation in pollen and nectar sources of honey bees in Ireland. Journal of Apicultural Research, 36(2), 63–76. https://doi.org/10.1080/00218839.1997.11100932
Cornman, R. S., Otto, C. R. V., Iwanowicz, D., & Pettis, J. S. (2015). Taxonomic characterization of honey bee (Apis mellifera) pollen foraging based on non-overlapping paired-end sequencing of nuclear ribosomal loci. PLoS ONE, 10, e0145365. https://doi.org/10.1371/journal.pone.0145365
Danner, N., Keller, A., Härtel, S., & Steffan-Dewenter, I. (2017). Honey bee foraging ecology: Season but not landscape diversity shapes the amount and diversity of collected pollen. PLoS ONE, 12(8), e0183716. https://doi.org/10.1371/journal.pone.0183716
Danner, N., Molitor, A. M., Schiele, S., Härtel, S., & Steffan-Dewenter, I. (2016). Season and landscape composition affect pollen foraging distances and habitat use of honey bees. Ecological Applications, 26(6), 1920–1929. https://doi.org/10.1890/15-1840.1
Delavaux, C. S., Bever, J. D., Karppinen, E. M., & Bainard, L. D. (2020). Keeping it cool: Soil sample cold pack storage and DNA shipment up to 1 month does not impact metabarcoding results. Ecology and Evolution, 10(11), 4652–4664. https://doi.org/10.1002/ece3.6219
Di Pasquale, G., Alaux, C., Le Conte, Y., Odoux, J.-F., Pioz, M., Vaissière, B. E., Belzunces, L. P., & Decourtye, A. (2016). Variations in the availability of pollen resources affect honey bee health. PLoS ONE, 11(9), e0162818. https://doi.org/10.1371/journal.pone.0162818
Di Pasquale, G., Salignon, M., Le Conte, Y., Belzunces, L. P., Decourtye, A., Kretzschmar, A., Suchail, S., Brunet, J.-L., & Alaux, C. (2013). Influence of pollen nutrition on honey bee health: Do pollen quality and diversity matter? PLoS ONE, 8(8), e72016. https://doi.org/10.1371/journal.pone.0072016
Dimou, M., & Thrasyvoulou, A. (2007). Seasonal variation in vegetation and pollen collected by honeybees in Thessaloniki. Greece. Grana, 46(4), 292–299. https://doi.org/10.1080/00173130701760718
Donkersley, P., Rhodes, G., Pickup, R. W., Jones, K. C., & Wilson, K. (2014). Honeybee nutrition is linked to landscape composition. Ecology and Evolution, 4(21), 4195–4206. https://doi.org/10.1002/ece3.1293
Doyle, J. J., & Dickson, E. E. (1987). Preservation of plant samples for DNA restriction endonuclease analysis. Taxon, 36(4), 715–722. https://doi.org/10.2307/1221122
Drummond, F. A., Ballman, E. S., Eitzer, B. D., Du Clos, B., & Dill, J. (2018). Exposure of honey bee (Apis mellifera L.) colonies to pesticides in pollen, a statewide assessment in Maine. Environmental Entomology, 47(2), 378–387. https://doi.org/10.1093/ee/nvy023
Dunker, S., Motivans, E., Rakosy, D., Boho, D., Mäder, P., Hornick, T., & Knight, T. M. (2021). Pollen analysis using multispectral imaging flow cytometry and deep learning. New Phytologist, 229(1), 593–606. https://doi.org/10.1111/nph.16882
Feinstein, L. M., Sul, W. J., & Blackwood, C. B. (2009). Assessment of bias associated with incomplete extraction of microbial DNA from soil. Applied and Environmental Microbiology, 75(16), 5428–5433. https://doi.org/10.1128/AEM.00120-09
Garbuzov, M., Couvillon, M. J., Schürch, R., & Ratnieks, F. L. W. (2015). Honey bee dance decoding and pollen-load analysis show limited foraging on spring-flowering oilseed rape, a potential source of neonicotinoid contamination. Agriculture, Ecosystems & Environment, 203, 62–68. https://doi.org/10.1016/j.agee.2014.12.009
Goulson, D., Nicholls, E., Botías, C., & Rotheray, E. L. (2015). Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science, 347(6229), 1255957. https://doi.org/10.1126/science.1255957
Gratzer, K., & Brodschneider, R. (2021). How and why beekeepers participate in the INSIGNIA citizen science honey bee environmental monitoring project. Environmental Science and Pollution Research. https://doi.org/10.1007/s11356-021-13379-7
Hawkins, J., de Vere, N., Griffith, A., Ford, C. R., Allainguillaume, J., Hegarty, M. J., Baillie, L., & Adams-Groom, B. (2015). Using DNA metabarcoding to identify the floral composition of honey: A new tool for investigating honey bee foraging preferences. PLoS ONE, 10(8), e0134735. https://doi.org/10.1371/journal.pone.0134735
Herbert, E. W., & Shimanuki, H. (1978). Chemical composition and nutritive value of bee-collected and bee-stored pollen. Apidologie, 9, 33–40. https://doi.org/10.1051/apido:19780103
Jones, L., Brennan, G. L., Lowe, A., Creer, S., Ford, C. R., & De Vere, N. (2021). Shifts in honeybee foraging reveal historical changes in floral resources. Communications Biology, 4(1), 1–10. https://doi.org/10.1038/s42003-020-01562-4
Kaluza, B. F., Wallace, H. M., Heard, T. A., Minden, V., Klein, A., & Leonhardt, S. D. (2018). Social bees are fitter in more biodiverse environments. Scientific Reports, 8(1), 12353. https://doi.org/10.1038/s41598-018-30126-0
Kandlikar, G. S., Gold, Z. J., Cowen, M. C., Meyer, R. S., Freise, A. C., Kraft, N. J., Moberg-Parker, J., Sprague, J., Kushner, D. J., & Curd, E. E. (2018). Ranacapa: an R package and shiny web app to explore environmental DNA data with exploratory statistics and interactive visualizations. F1000Research, 7, 1734. https://doi.org/10.12688/f1000research.16680.1
Keller, A., Danner, N., Grimmer, G., Ankenbrand, M., Von Der Ohe, K., Von Der Ohe, W., Rost, S., Härtel, S., & Steffan-Dewenter, I. (2015). Evaluating multiplexed next-generation sequencing as a method in palynology for mixed pollen samples. Plant Biology, 17, 558–566. https://doi.org/10.1111/plb.12251
Kindt, R., & Coe, R. (2005). Tree diversity analysis: a manual and software for common statistical methods for ecological and biodiversity studies. Nairobi: World Agroforestry Centre (ICRAF).
Koffler, S., Barbiéri, C., Ghilardi-Lopes, N. P., Leocadio, J. N., Albertini, B., Francoy, T. M., & Saraiva, A. M. (2021). A buzz for sustainability and conservation: The growing potential of citizen science studies on bees. Sustainability, 13(2), 959. https://doi.org/10.3390/su13020959
Kraaijeveld, K., De Weger, L. A., Ventayol García, M., Buermans, H., Frank, J., Hiemstra, P. S., & Den Dunnen, J. T. (2015). Efficient and sensitive identification and quantification of airborne pollen using next-generation DNA sequencing. Molecular Ecology Resources, 15, 8–16. https://doi.org/10.1111/1755-0998.12288
Lau, P., Bryant, V., Ellis, J. D., Huang, Z. Y., Sullivan, J., Schmehl, D. R., Cabrera, A. R., & Rangel, J. (2019). Seasonal variation of pollen collected by honey bees (Apis mellifera) in developed areas across four regions in the United States. PLoS ONE, 14(6), e0217294. https://doi.org/10.1371/journal.pone.0217294
Liu, M., Clarke, L. J., Baker, S. C., Jordan, G. J., & Burridge, C. P. (2020). A practical guide to DNA metabarcoding for entomological ecologists. Ecological Entomology, 45(3), 373–385. https://doi.org/10.1111/een.12831
Macgregor, C. J., Kitson, J. J. N., Fox, R., Hahn, C., Lunt, D. H., Pocock, M. J. O., & Evans, D. M. (2019). Construction, validation, and application of nocturnal pollen transport networks in an agro-ecosystem: A comparison using light microscopy and DNA metabarcoding. Ecological Entomology, 44(1), 17–29. https://doi.org/10.1111/een.12674
McMurdie, P. J., & Holmes, S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE, 8, e61217. https://doi.org/10.1371/journal.pone.0061217
Milla, L., Sniderman, K., Lines, R., Mousavi-Derazmahalleh, M., & Encinas-Viso, F. (2021). Pollen DNA metabarcoding identifies regional provenance and high plant diversity in Australian honey. Ecology and Evolution, 11, 8683–8698. https://doi.org/10.1002/ece3.7679
Miller-Rushing, A., Primack, R., & Bonney, R. (2012). The history of public participation in ecological research. Frontiers in Ecology and the Environment, 10(6), 285–290. https://doi.org/10.1890/110278
Moro, A., Beaurepaire, A., Dall’Olio, R., Rogenstein, S., Blacquière, T., Dahle, B., de Miranda, J. R., Dietemann, V., Locke, B., Licón Luna, R. M., Le Conte, Y., & Neumann, P. (2021). Using citizen science to scout honey bee colonies that naturally survive Varroa destructor infestations. Insects, 12(6). https://doi.org/10.3390/insects12060536
Murray, M. G., & Pitas, J. W. (1996). Plant DNA from alcohol-preserved samples. Plant Molecular Biology Reporter, 14(3), 261–265. https://doi.org/10.1007/BF02671661
Nagy, Z. T. (2010). A hands-on overview of tissue preservation methods for molecular genetic analyses. Organisms Diversity & Evolution, 10(1), 91–105. https://doi.org/10.1007/s13127-010-0012-4
Nürnberger, F., Keller, A., Härtel, S., & Steffan-Dewenter, I. (2019). Honey bee waggle dance communication increases diversity of pollen diets in intensively managed agricultural landscapes. Molecular Ecology, 28(15), 3602–3611. https://doi.org/10.1111/mec.15156
Ogle, D. H., Doll, J. D., Wheeler, P., & Dinno, A. (2021). FSA: Fisheries Stock Analysis. R package version 0.9.0.9000, 636. https://github.com/droglenc/FSA
Omar, A., Kadesjö, N., Palmgren, C., Marteinsdottir, M., Segerdahl, T., & Fransson, A. (2017). Assessment of the occupational eye lens dose for clinical staff in interventional radiology, cardiology and neuroradiology. Journal of Radiological Protection, 37(1), 145–159. https://doi.org/10.1088/1361-6498/aa559c
Pacini, E., & Hesse, M. (2005). Pollenkitt – its composition, forms and functions. Flora - Morphology, Distribution, Functional Ecology of Plants, 200(5), 399–415. https://doi.org/10.1016/j.flora.2005.02.006
Pawluczyk, M., Weiss, J., Links, M. G., Egaña Aranguren, M., Wilkinson, M. D., & Egea-Cortines, M. (2015). Quantitative evaluation of bias in PCR amplification and next-generation sequencing derived from metabarcoding samples. Analytical and Bioanalytical Chemistry, 407(7), 1841–1848. https://doi.org/10.1007/s00216-014-8435-y
Piñol, J., Senar, M. A., & Symondson, W. O. C. (2019). The choice of universal primers and the characteristics of the species mixture determine when DNA metabarcoding can be quantitative. Molecular Ecology, 28(2), 407–419. https://doi.org/10.1111/mec.14776
Pompanon, F., Deagle, B. E., Symondson, W. O. C., Brown, D. S., Jarman, S. N., & Taberlet, P. (2012). Who is eating what: Diet assessment using next generation sequencing. Molecular Ecology, 21(8), 1931–1950. https://doi.org/10.1111/j.1365-294X.2011.05403.x
Pornon, A., Escaravage, N., Burrus, M., Holota, H., Khimoun, A., Mariette, J., Pellizzari, C., Iribar, A., Etienne, R., Taberlet, P., Vidal, M., Winterton, P., Zinger, L., & Andalo, C. (2016). Using metabarcoding to reveal and quantify plant-pollinator interactions. Scientific Reports, 6(1), 27282. https://doi.org/10.1038/srep27282
Potter, C., de Vere, N., Jones, L. E., Ford, C. R., Hegarty, M. J., Hodder, K. H., Diaz, A., & Franklin, E. L. (2019). Pollen metabarcoding reveals broad and species-specific resource use by urban bees. PeerJ, 7, e5999. https://doi.org/10.7717/peerj.5999
Prendini, L., Hanner, R., & DeSalle, R. (2002). Obtaining, storing and archiving specimens and tissue samples for use in molecular studies. In R. DeSalle, G. Giribet, & W. Wheeler (Eds.), Techniques in Molecular Systematics and Evolution (pp. 176–248). Birkhäuser Basel. https://doi.org/10.1007/978-3-0348-8125-8_11
Rasmussen, C., Dupont, Y. L., Madsen, H. B., Bogusch, P., Goulson, D., Herbertsson, L., Maia, K. P., Nielsen, A., Olesen, J. M., Potts, S. G., Roberts, S. P. M., Sydenham, M. A. K., & Kryger, P. (2021). Evaluating competition for forage plants between honey bees and wild bees in Denmark. PLoS ONE, 16(4), e0250056. https://doi.org/10.1371/journal.pone.0250056
Requier, F., Odoux, J.-F., Tamic, T., Moreau, N., Henry, M., Decourtye, A., & Bretagnolle, V. (2015). Honey bee diet in intensive farmland habitats reveals an unexpectedly high flower richness and a major role of weeds. Ecological Applications, 25(4), 881–890. https://doi.org/10.1890/14-1011.1
Richardson, R. T., Bengtsson-Palme, J., & Johnson, R. M. (2017). Evaluating and optimizing the performance of software commonly used for the taxonomic classification of DNA metabarcoding sequence data. Molecular Ecology Resources, 17(4), 760–769. https://doi.org/10.1111/1755-0998.12628
Richardson, R. T., Eaton, T. D., Lin, C.-H., Cherry, G., Johnson, R. M., & Sponsler, D. B. (2019). Honey bee pollen foraging ecology across an urbanization gradient. bioRxiv, 824474. https://doi.org/10.1101/824474
Richardson, R. T., Lin, C.-H., Quijia, J. O., Riusech, N. S., Goodell, K., & Johnson, R. M. (2015a). Rank-based characterization of pollen assemblages collected by honey bees using a multi-locus metabarcoding approach. Applications in Plant Sciences, 3(11), 1500043. https://doi.org/10.3732/apps.1500043
Richardson, R. T., Lin, C. H., Sponsler, D. B., Quijia, J. O., Goodell, K., & Johnson, R. M. (2015b). Application of ITS2 metabarcoding to determine the provenance of pollen collected by honey bees in an agroecosystem. Applications in Plant Sciences, 3, 1400066. https://doi.org/10.3732/apps.1400066
Rognes, T., Flouri, T., Nichols, B., Quince, C., & Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ, 4, e2584. https://doi.org/10.7717/peerj.2584
Rubin, B. E. R., Gibbons, S. M., Kennedy, S., Hampton-Marcell, J., Owens, S., & Gilbert, J. A. (2013). Investigating the impact of storage conditions on microbial community composition in soil samples. PLoS ONE, 8(7), e70460. https://doi.org/10.1371/journal.pone.0070460
Schiebelhut, L. M., Abboud, S. S., Gómez Daglio, L. E., Swift, H. F., & Dawson, M. N. (2017). A comparison of DNA extraction methods for high-throughput DNA analyses. Molecular Ecology Resources, 17(4), 721–729. https://doi.org/10.1111/1755-0998.12620
Sickel, W., Ankenbrand, M. J., Grimmer, G., Holzschuh, A., Härtel, S., Lanzen, J., Steffan-Dewenter, I., & Keller, A. (2015). Increased efficiency in identifying mixed pollen samples by meta-barcoding with a dual-indexing approach. BMC Ecology, 15(1), 1–9. https://doi.org/10.1186/s12898-015-0051-y
Smart, M. D., Cornman, R. S., Iwanowicz, D. D., McDermott-Kubeczko, M., Pettis, J. S., Spivak, M. S., & Otto, C. R. V. (2017). A comparison of honey bee-collected pollen from working agricultural lands using light microscopy and ITS metabarcoding. Environmental Entomology, 46(1), 38–49. https://doi.org/10.1093/ee/nvw159
Smart, M. D., Pettis, J. S., Euliss, N., & Spivak, M. S. (2016). Land use in the Northern Great Plains region of the U.S. influences the survival and productivity of honey bee colonies. Agriculture, Ecosystems & Environment, 230, 139–149. https://doi.org/10.1016/j.agee.2016.05.030
Swenson, S. J., & Gemeinholzer, B. (2021). Testing the effect of pollen exine rupture on metabarcoding with Illumina sequencing. PLoS ONE, 16(2), e0245611. https://doi.org/10.1371/journal.pone.0245611
Team, R. (2015). RStudio: integrated development for R. In RStudio Inc. http://www.rstudio.com
Tosi, S., Costa, C., Vesco, U., Quaglia, G., & Guido, G. (2018). A 3-year survey of Italian honey bee-collected pollen reveals widespread contamination by agricultural pesticides. Science of the Total Environment, 615, 208–218. https://doi.org/10.1016/j.scitotenv.2017.09.226
Weißbecker, C., Buscot, F., & Wubet, T. (2017). Preservation of nucleic acids by freeze-drying for next generation sequencing analyses of soil microbial communities. Journal of Plant Ecology, 10(1), 81–90. https://doi.org/10.1093/jpe/rtw042
White, T. J., Bruns, T., Lee, S. J. W. T., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In G. D. M. A Innis, J. J. Sninsky, & T. J. White (Eds.), PCR protocols: a guide to methods applications (pp. 315–322). Academic Press
Acknowledgements
We are deeply indebted to Susana Lopes and Maria Magalhães, from CIBIO—Research Centre in Biodiversity and Genetic Resources—InBIO Associate Laboratory, for their time devoted to library preparation and sequencing in the MiSeq. AQ acknowledges the PhD scholarship (DFA/BD/5155/2020) funded by FCT.
Funding
This work was funded by the Health and Food Safety Directorate General, European Commission through the project INSIGNIA—Environmental monitoring of pesticide use through honeybees SANTE/E4/SI2.788418-SI2.788452-INSIGINIA-PP-1–1-2018. Fundação para a Ciência e a Tecnologia (FCT) provided financial support by national funds (FCT/MCTES) to CIMO (UIDB/00690/2020).
Author information
Authors and Affiliations
Contributions
MAP, JvdS, RB, OK and FV conceived the ideas and designed the methodology. AQ performed the laboratorial metabarcoding work as well as all the analyses with assistance of MAP and AG. FV and OK performed the preliminary silica testing. Pollen sampling and sample preservation were carried out by RB, KG, OK and FV. AK and JR assembled the reference ITS2 database and assisted with bioinformatics. JvdS acquired funding. MAP and AQ wrote the original draft. All the authors contributed with data interpretation. All the authors critically revised and edited the manuscript for important intellectual content.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Quaresma, A., Brodschneider, R., Gratzer, K. et al. Preservation methods of honey bee-collected pollen are not a source of bias in ITS2 metabarcoding. Environ Monit Assess 193, 785 (2021). https://doi.org/10.1007/s10661-021-09563-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-021-09563-4