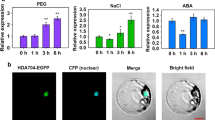
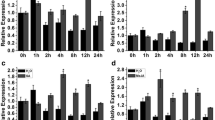
Abstract
Acetylation and deacetylation of histones are important for regulating a series of biological processes in plants. Histone deacetylases (HDACs) control the histone deacetylation that plays an important role in plant response to abiotic stress. In our study, we show the evidence that GhHDT4D (a member of the HD2 subfamily of HDACs) is involved in cotton (Gossypium hirsutum) response to drought stress. Overexpression of GhHDT4D in Arabidopsis increased plant tolerance to drought, whereas silencing GhHDT4D in cotton resulted in plant sensitivity to drought. Simultaneously, the H3K9 acetylation level was altered in the GhHDT4D silenced cotton, compared with the controls. Further study revealed that GhHDT4D suppressed the transcription of GhWRKY33, which plays a negative role in cotton defense to drought, by reducing its H3K9 acetylation level. The expressions of the stress-related genes, such as GhDREB2A, GhDREB2C, GhSOS2, GhRD20-1, GhRD20-2 and GhRD29A, were significantly decreased in the GhHDT4D silenced cotton, but increased in the GhWRKY33 silenced cotton. Given these data together, our findings suggested that GhHDT4D may enhance drought tolerance by suppressing the expression of GhWRKY33, thereby activating the downstream drought response genes in cotton.







Similar content being viewed by others
References
Aubert Y, Vile D, Pervent M, Aldon D, Ranty B, Simonneau T, Galaud J (2010) RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol 51:1975–1987
Annunziato AT, Hansen JC (2001) Role of histone acetylation in the assembly and modulation of chromatin structures. Gene expression. J Liver Res 9(1–2):37–61
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43:W39–W49
Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146:368–376
Chen J, Nolan TM, Ye H, Zhang M, Tong H, Xin P, Yin Y (2017) Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29:1425–1439
Chen LT, Wu K (2010) Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal Behav 5:1318–1320
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys A 1819:120–128
Chen C, Chen H, He Y, Xia R (2018) TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv. https://doi.org/10.1101/289660
Chen Y, Liu ZH, Feng L, Zheng Y, Li DD, Li XB (2013) Genome-wide functional analysis of cotton (Gossypium hirsutum) in response to drought. PLoS ONE 8(11):120–128
Chen HP, Zhao YT, Zhao TC (2015) Histone deacetylases and mechanisms of regulation of gene expression. Crit Rev Oncog. 20:35–47
Chrun ES, Modolo F, Daniel FI (2018) HDAC1 (histone deacetylase 1). Atlas of genetics and cytogenetics in oncology and haematology.
Choi SM, Song HR, Han SK, Han M, Kim CY, Park JJ, Noh B (2012) HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J 71:135–146
De Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Hulo N (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34(2):W362–W365
Farhi JA (2015) HD2D is a regulator of abscisic acid responses in Arabidopsis. Electronic thesis and dissertation repository 3467.
Gao W, Long L, Xu L, Lindsey K, Zhang X, Zhu L (2016) Suppression of the homeobox gene HDTF1 enhances resistance to Verticillium dahliae and Botrytis cinerea in cotton. J Integr Plant Biol 58:503–513
Gotmare V, Singh P, Mayee CD, Deshpande V, Bhagat C (2004) Genetic variability for seed oil content and seed index in some wild species and perennial races of cotton. Plant Breed 123:207–208
Han Z, Yu H, Zhao Z, Hunter D, Luo X, Duan J, Tian L (2016) AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in Arabidopsis thaliana. Front Plant Sci 7:310
Hollender C, Liu Z (2008) Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol 50:875–885
Je J, Chen H, Song C, Lim CO (2014) Arabidopsis DREB2C modulates ABA biosynthesis during germination. Biochem Biophys Res Commun 452:91–98
Kim JM, Sasaki T, Ueda M, Sako K, Seki M (2015) Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Frontiers Plant Sci 6:114
Koprinarova M, Schnekenburger M, Diederich M (2016) Role of histone acetylation in cell cycle regulation. Curr Top Med Chem 16(7):732–744
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287
Legube G, Trouche D (2003) Regulating histone acetyltransferases and deacetylases. EMBO Rep 4:944–947
Liu X, Yang S, Zhao M, Luo M, Yu CW, Chen CY, Wu K (2014) Transcriptional repression by histone deacetylases in plants. Mol Plant 7:764–772
Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97:3730–3734
Liu ZH, Chen Y, Wang NN, Chen YH, Wei N, Lu R, Li XB (2019) A basic helix—loop—helix (bHLH) protein (GhFP1) promotes fiber elongation of cotton (Gossypium hirsutum) via modulating BR biosynthesis and signaling. New Phytol. https://doi.org/10.1111/nph.16301
Luo M, Liu X, Singh P, Cui Y, Zimmerli L, Wu K (2012a) Chromatin modifications and remodeling in plant abiotic stress responses. Biochim Biophys Acta 1819:129–136
Luo M, Wang YY, Liu X, Yang S, Lu Q, Cui Y, Wu K (2012b) HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 63:3297–3306
Luo M, Wang YY, Liu X, Yang S, Wu K (2012c) HD2 proteins interact with RPD3-type histone deacetylases. Plant Signal Behav 7:608–610
Luo M, Cheng K, Xu Y, Yang S, Wu K (2017) Plant responses to abiotic stress regulated by histone deacetylases. Frontiers Plant Sci 8:2147
Ma X, Lv S, Zhang C, Yang C (2013) Histone deacetylases and their functions in plants. Plant Cell Rep 32(4):465–478
Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2000) Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration-and high-salinity-responsive gene expression. Plant Mol Biol 42:657–665
Perrella G, Lopez-Vernaza MA, Carr C, Sani E, Gosselé V, Verduyn C (2013) Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25:3491–3505
Peterson CL, Laniel MA (2004) Histones and histone modifications. Curr Biol 14:R546–R551
Pettigrew WT (2004) Physiological consequences of moisture deficit stress in cotton. Crop Sci 44:1265–1272
Pfluger J, Wagner D (2007) Histone modifications and dynamic regulation of genome accessibility in plants. Curr Opin Plant Biol 10:645–652
Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18:1292–1309
Song J, Henry HA, Tian L (2019) Brachypodium histone deacetylase BdHD1 positively regulates ABA and drought stress responses. Plant Sci 283:355–365
Sridha S, Wu K (2006) Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J 46(1):124–133
Wai AH, An G (2018) Histone Deacetylase 701 enhances abiotic stress resistance in rice at the seedling stage by suppressing expression of OsWRKY45. 2nd Myanmar-Korea conference on useful plants, Dagon University.
Wang C, Gao F, Wu J, Dai J, Wei C, Li Y (2010) Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol 51(8):1291–1299
Wang NN, Xu SW, Sun YL, Liu D, Zhou L, Li Y, Li XB (2019) The cotton WRKY transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Sci Rep 9:724
Xiao BZ, Chen X, Xiang CB, Tang N, Zhang QF, Xiong LZ (2009) Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol Plant 2:73–83
Xiu Y, Iqbal A, Zhu C, Wu G, Chang Y, Li N, Wang H (2016) Improvement and transcriptome analysis of root architecture by overexpression of Fraxinus pennsylvanica DREB2A transcription factor in Robinia pseudoacacia L. ‘Idaho’. Plant Biotechnol J 14:1456–1469
Yao Y, Yang YW, Liu JY (2006) An efficient protein preparation for proteomic analysis of developing cotton fibers by 2-DE. Electrophoresis 27:4559–4569
Zhang F, Wang L, Ko EE, Shao K, Qiao H (2018) Histone deacetylases SRT1 and SRT2 interact with ENAP1 to mediate ethylene-induced transcriptional repression. Plant Cell 30(1):153–166
Zhao J, Zhang J, Zhang W, Wu K, Zheng F, Tian L, Duan J (2015) Expression and functional analysis of the plant-specific histone deacetylase HDT701 in rice. Front Plant Sci 5:764
Zhao K, Shen X, Yuan H, Liu Y, Liao X, Wang Q, Li T (2013) Isolation and characterization of dehydration-responsive element-binding factor 2C (MsDREB2C) from Malus sieversii Roem. Plant Cell Physiol 54:1415–1430
Zheng Y, Ding Y, Sun X, Xie S, Wang D, Liu X, Zhou DX (2016) Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J Exp Bot 67:1703–1713
Zhou C, Labbe H, Sridha S, Wang L, Tian L, Latoszek-Green M, Wu K (2004) Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J 38(5):715–724
Zhou DX, Hu Y (2010) Regulatory function of histone modifications in controlling rice gene expression and plant growth. Rice 3(2):103–111
Zhou J, Wang X, He K, Charron JBF, Elling AA, Deng XW (2010) Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol Biol 72:585–595
Zhu G, Li W, Zhang F, Guo W (2018) RNA-seq analysis reveals alternative splicing under salt stress in cotton, Gossypium davidsonii. BMC Genom 19(1):73
Acknowledgements
This work was financially supported by National Key R&D Program of China (Grant No. 2016YFD0100505), and National Natural Sciences Foundation of China (Grant No. 31871667).
Author information
Authors and Affiliations
Contributions
XBL and JBZ conceived and designed the research; JBZ, SPH, JWL, XPW and DDL performed the experiments; JBZ and XBL analyzed data and wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no any competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, JB., He, SP., Luo, JW. et al. A histone deacetylase, GhHDT4D, is positively involved in cotton response to drought stress. Plant Mol Biol 104, 67–79 (2020). https://doi.org/10.1007/s11103-020-01024-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01024-9