Abstract
Amyloidogenesis is the inherent ability of proteins to change their conformation from native state to cross β-sheet rich fibrillar structures called amyloids which result in a wide range of diseases like Parkinson's disease, Alzheimer’s disease, Finnish familial amyloidosis, ATTR amyloidosis, British and Danish dementia, etc. COVID-19, on the other hand is seen to have many similarities in symptoms with other amyloidogenic diseases and the overlap of these morbidities and symptoms led to the proposition whether SARS-CoV-2 proteins are undergoing amyloidogenesis and whether it is resulting in or aggravating amyloidogenesis of any human host protein. Thus the SARS-CoV-2 proteins in infected cells, i.e., Spike (S) protein, Nucleocapsid (N) protein, and Envelope (E) protein were tested via different machinery and amyloidogenesis in them were proven. In this review, we will analyze the pathway of amyloid formation in S-protein, N-protein, E-protein along with the effect that SARS-CoV-2 is creating on various host proteins leading to the unexpected onset of many morbidities like COVID-induced Acute Respiratory Distress Syndrome (ARDS), Parkinsonism in young COVID patients, formation of fibrin microthrombi in heart, etc., and their future implications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Protein folding and binding provide the basis for life on earth. The native 3D structure of a protein is necessary for its biological function (Perozzo et al. 2004). Among many protein folding models such as diffusion-collision, nucleation-condensation, jigsaw puzzle, hydrophobic collapse and stoichiometry models, the “folding funnel” model based on the free energy landscape theory has now been most widely accepted (Shu-Qun Liu et al. 2012). The natural tendency of polypeptide chains to get folded into its unique native structure inside a cell is guided by many machinery like the proteasomes, chaperones, protein disulfide isomerases, ubiquitins, prolyl peptide isomerase, etc., which reserve the properly folded proteins and degrade the misfolded ones. Though there are a lot of corrective machineries present, yet proteins have an inherent tendency to undergo transition to self-assembled aggregates from their native soluble state. Of the various types of protein aggregates, “amyloids” are a type of stable, fibrillar, ordered protein aggregates which are possess cross β sheet-rich structures and the process of amyloid formation is called amyloidogenesis (Dobson 1999; Clark et al. 1981). Till date, about 37 human proteins have been seen to form amyloids and most of them are related to several degenerative disorders like Parkinson’s disease, Alzheimer’s disease, type II diabetes, etc., (Marzban et al. 2003; Ghetti et al. 1996; Ghiso and Frangione 2002; Petrou et al. 2015). Apart from pathogenicity, amyloids have been also found to be functional in many aspects, as structural components in bacteria and viruses, as biochemical regulators functioning as hemostatic agent in human beings, as scaffolds, molecular chaperones as well as in sexual reproduction (Sarkar 2020).
Now, coming to the current scenario, the world has been almost put to a standstill in the post-pandemic era as we are still recovering from the damage it has caused us, be it physically, mentally or economically. With the intensive research going on SARS-CoV-2, scientists noticed similarities in symptoms and morbidities between COVID-19 and amyloid regulated diseases, which led to the hypothetical link of amyloidogenesis in coronavirus. Supported by the prior proof about interactions between amyloids present in viruses and their respective human host, various research work was conducted which brought forward the link between COVID-19 and amyloid regulated diseases like Parkinson’s, Alzheimer's, ATTR (amyloidogenesis of transthyretin), etc., (Tayeb-Fligelman et al. 2021; Michiels et al. 2021; Munch et al. 2007; Dimitrov et al. 1993). This review article presents a summary of almost all the current work done on amyloidogenesis in coronavirus, and how that may aggravate amyloidosis in future.
Amyloidogenesis in SARS-CoV-2 proteins
Similarities in morbidities between COVID-19 and amyloidogenic cardiopathy and neuropathy like heart failure, blood clotting, CNS disorders, peripheral neuropathy, etc., and unusual onset of what is known as “Parkinsonism” which include conditions related to abnormalities in movement as seen in Parkinson's Disease, post-COVID recovery in young patients led to the hypothesis that maybe different proteins of SARS-CoV-2 are amyloidogenic, based on the prior proof of amyloidogenesis in viral proteins that infect human systems like the liver, kidney, immune cells and even the CNS (Merello et al. 2021; Cohen et al. 2020; Lee et al. 2021).
SARS-CoV, the coronavirus responsible for the outbreak of SARS in 2003 has been experimentally proven to contain amyloidogenic proteins and since the proteome of SARS-CoV and SARS-CoV-2 have many similarities in biological structure and function, it was proposed that SARS-CoV-2 also contains amyloidogenic proteins which can give rise to neurodegenerative complications (Rangan et al. 2020; Galkin 2021). In a study, open reading frames (ORFs) of SARS-CoV-2 proteome were studied using a computational tool named ZIPPER for screening amyloidogenic sequences which led to the result two sub-sequences from ORF6 and ORF10 were aggregation prone (Charnley et al. 2022). The main structural and functional proteins including the non-structural proteins (NSPs) of SARS-CoV and SARS-CoV-2 was computationally screened using four softwares namely MetAmyl, FISH Amyloid, AGGRESCAN, and FoldAmyloid to find out aggregation prone regions (APRs) in both the proteomes. In the proteome of SARS-CoV, the membrane protein or M-protein, C-terminal end and transmembrane domain (TMD) of envelope protein or E-protein, ORF8b have been proven to be amyloidogenic (Ghosh et al. 2015; Lee et al. 2005). In SARS-CoV-2, membrane protein (M), envelope protein (E), among the structural proteins along with the accessory proteins were found to be more amyloidogenic than nucleocapsid protein (N) and spike protein (S). Out of the 16 non-structural proteins (NSPs) present in the genome of SARS-CoV-2, NSP4 and NSP6 were found to be highly amyloidogenic. Besides, mean predicted percentage amyloidogenic propensity study revealed that accessory proteins of SARS-CoV-2 were more aggregation prone than that of SARS-CoV (Bhardwaj et al. 2021).
Spike protein (S-protein) is the primary SARS-CoV-2 contact protein between the host and the virus which helps in virus docking and host-entry (Nyström and Hammarström 2021). Nucleocapsid protein (N-protein) on the other hand is more abundant, relatively more stable and conserved than the S-protein gene (Cubuk et al. 2021). Envelope protein (E protein), though being the smallest, interacts with other proteins and helps in maintaining the viral shape, release and also promotes cellular apoptosis (Alsaadi et al. 2020; Chen et al. 2009). All these proteins were tested through various experiments to evaluate their amyloidogenic propensity, and the results are demonstrated in Fig. 1. It was furthermore found out through computational study that the above mentioned proteins in SARS-CoV-2 have more aggregation tendency than those of SARS-CoV-2 (Bhardwaj et al. 2021).
Graphical representation of the SARS-CoV-2 protein undergoing/accelerating amyloidogenesis; A S-protein digested by serine protease enzyme like neutrophil esterase results in formation of amyloid-prone segment 193–202 which later forms amyloids; B N-protein (left) interacts with amyloidogenic α-Synuclein protein (right) and results in acceleration of amyloidogenesis; C E-protein destabilizes by hydrophobic interaction with the environment into nine-residue segment TK9 which forms amyloids by self-assembly
SARS-CoV-2 Spike Protein Amyloidogenesis:
Complete S-protein sequence (ID: P0DTC2) was subjected to invitro fibril formation and seven amyloidogenic sequences were deciphered out of several 20 amino acid long sequences, using the WALTZ algorithm, which were named according to the start position of the S‐protein; S191, S-259, S-362, S-532, S-599, S-689, S-1165, as shown in Table 1. Out of these, S-362 was seen to not have the cross β-sheet conformation under cryo-EM. In vitro amyloidogenesis study of lyophilized peptide using ThT formation kinetics, Congo-Red birefringence (CR) and transmission electron microscopy (TEM) showed S-191, S-532 and S-1165 to fulfill all the necessary amyloid criteria, out of which S-191 showed the most ordered fibrils (Zhang et al. 2018; Maurer-Stroh et al. 2010; Hamodrakas et al. 2007; Galzitskaya et al. 2006). Furthermore, sigmoidal kinetics curve also predicted dominance of S-191 fibrils. Thus, out of all the assumed amyloidogenic sequences, spike peptide S-191 showed most potent amyloidogenicity fulfilling almost all the authenticity criteria, proving the existence of amyloidogenesis in S-protein of SARS-CoV-2. Owing to the high stability (Tm > 50) and complex fold structure, SARS-CoV-2 S-protein is not spontaneous to amyloid formation (Upadhyay et al. 2021). Since endoproteolysis of amyloid prone proteins or even full length proteins leads to the molecular initiation of many amyloidogenic diseases like Alzheimer's, Finnish familial amyloidosis, British and Danish dementia, ATTR amyloidosis, etc., it was considered as a hypothetical mechanism to start with, which was further experimentally investigated (Sipe 2008). SARS-CoV-2 S-protein is proteolysed during infection and also during inflammation by host furin-like enzymes and by release of enzymes like neutrophil esterase (abbreviated as NE, a serine protease that causes obstructive lung diseases such as COPD, cystic fibrosis and alpha‐1‐antitrypsin deficiency) by the neutrophils extracellularly, which are recruited by the host immune system to the bronchoalveolar cavity of patients affected with various respiratory viruses, including SARS‐CoV‐2 (Peacock et al. 2021; Johansson and Kirsebom 2021; Strnad et al. 2020). At first, in-silico proteolytic cleavage of full length S-protein sequence by NE was done using Expasy Peptide cutter and one of the peptides from the results, Spike 193–212 matched with S-191, only with a frame shift by 2 amino acids. This proved the hypothetical mechanism and thus was subjected to further in vitro testing by S-protein digestion with NE. Among all the peptide segments formed segment 193‐202 (FKNIDGYFKI, included in Spike191) was highly abundant after 6 h of incubation, which made it amyloidogenically important (Nyström and Hammarström 2021).
Acceleration of amyloidogenesis due to SARS-CoV-2 N-Protein:
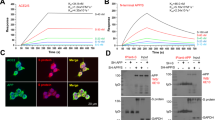
Reports of unexpected Parkinsonism in young patients after recovering from SARS-CoV-2 infection raised the question whether any SARS-CoV-2 protein is leading to accelerated amyloidogenesis of α-synuclein (αS) protein that leads to the formation of Parkinson’s Disease (PD) (Espay and Henderson 2011; Bantle et al. 2019; Fishman et al. 1985; Merello et al. 2021; Cohen et al. 2020). N-protein being more stable and conserved, was chosen for investigation of the above-mentioned hypothesis. As shown in Fig. 2, ThT assay of αS peptide in the presence of N-protein resulted in reduction in aggregation lag time to less than 24 h, which decreased with the increase in N-protein concentration. This clearly indicated that the presence of N-protein was considerably accelerating the amyloidogenesis in αS-protein. Since, at a near neutral pH of 7.4, N-protein is positively charged (+ 24e) whereas αS is negatively charged (-9e), thus, electrostatic attraction is thought to be the primary intermolecular interactive force (Semerdzhiev et al. 2022; Taquet et al. 2021). Further in another study, microscale thermophoresis (MST) and fluorescence correlation spectroscopy (FCS) assay was performed on fluorescently labeled αS and with increasing concentration of the N-protein which predicted the presence of about 3 to 4 αS proteins in an αS/N-protein complex along with the indication of cooperative binding (Chatterjee et al. 2021).
Aggregation of αS in the absence and presence of SARS-CoV-2 proteins. a Aggregation assay of αS in the absence (black) and presence (color) of S-protein. The aggregation process is followed by recording the fluorescence of the amyloid-binding dye ThT. The assay was performed at a NaCl concentration of 10 mM with 50 μM αS and 0.1 μM (red), 0.5 μM (orange), and 1 μM (blue) S-protein. b ThT-based aggregation assay of αS in the presence of N-protein. The assay was performed at a salt concentration of 10 mM NaCl with 50 μM αS and 0 μM (black) 0.1 μM (red), 0.5 μM (orange), 0.8 μM (green) and 1 μM (blue) N-protein. c Influence of the salt concentration on aggregation lag time for N-protein concentrations of 0.5 μM (orange), 0.8 μM (green) and 1 μM (blue) at an αS concentration of 50 μM. The points represent the mean of three independent measurements, and error bars show the standard deviation. Figure taken with permission from Ref. (Semerdzhiev et al. 2022)
While the S-protein of SARS-CoV-2 is responsible for host entry, the N-protein is predominantly found in the cytoplasm post-infection, of the order of about 500 nM (Chang et al. 2004; Bar-On et al. 2020; Timani et al. 2005). Thus, to investigate the effect of N-protein presence along with αS in the cellular environment, microinjection of appropriate amount of N-protein as present during infection was done in SH-SY5Y neuronal cell model, which express αS peptide and extensively used in PD research. Now, the endogenously disordered αS is bound to vesicles, which take part in membrane remodeling and in membrane trafficking processes having an α-helical conformation that can be differentiated from the unbound ones (Fakhree et al. 2016; Kaur and Lee 2020; Burré et al. 2014; Lautenschläger et al. 2017). Förster resonance energy transfer (FRET) probes were used in vitro to detect these conformational changes and locate such vesicle bound αS in cells. The FRET results showed that compared to the control where only FRET-labeled αS peptides were present, high FRET signals were less indicating that the presence of N-protein decelerates the endogenous αS proteostasis, ultimately reducing the number of vesicle-bound αS (Fakhree et al. 2018; Nemani et al. 2010). The control probe was prepared by attaching the FRET-probe with the N-terminal domain of αS peptide and it did not show any change in FRET readings, indicating that αS is likely to bind to the C-terminal region of N-protein. These studies proved the involvement of SARS-CoV-2 N-protein in amyloidogenesis of αS peptide through direct molecular contact that leads to proteostasis of the peptide and hampers its normal cellular functioning, thereby inducing Parkinsonism in many unusual cases, post infection.
Amyloidogenesis in SARS-CoV-2 E-protein:
SARS-CoV-2 E protein has been said to have role imparting virulence and has been proved to be responsible for transferring the other corona proteins to the Golgi complex for further infective modifications. A β-sheet conformatory region, 55-TVYVYSRVK-63 (TK9), which contains residues that have similarities with many short length amyloid proteins like amyloid-β, is considered to be critical for this function (Li et al. 2014; Halverson et al. 1990). This led to the hypothesis that maybe TK9 is amyloidogenic which may result in enhanced virulence of SARS-CoV-2 strain. Now, in short length amyloidogenic peptides self-assembly has been seen as a potential mechanism of protein misfolding and thus the self-assembling potential of the nine-residue peptide sequence TK9 was tested (Lu et al. 2003). Dynamic light scattering (DLS), circular dichroism (CD) and ThT assay studies showed that proper β-sheet type spectral signature was seen after about 15 days of incubation, henceforth indicative of the fact that amyloidogenic propensity increases with time in E-protein (Ghosh et al. 2015; Lawrence et al. 1995). Another study determined that the probable mechanism behind the self-assembling nature of TK9 is that due to increase in hydrophobic and aromatic residues in the environment, hydrophobic bonding as well as π − π interactions increases between the amyloid aggregation-prone motifs of peptidemers which result in steady transition to cross β-sheet nature by self-assembly, resulting in the formation of amyloids. Moreover these motifs can easily bind with other amyloid prone proteins in the host and result in acceleration of amyloidogenesis of those proteins leading to some peculiar morbidity (Lopez de la Paz and Serrano 2004; Minor and Kim 1994).
Amyloidogenesis in human-host proteins as a result of COVID-19
COVID-19, though being a respiratory system infection, has a plethora of other symptoms which spread almost to every other system in the body ranging from neurological/sensory problems like loss of taste and smell, fatigue; gastrointestinal problems like nausea, diarrhea; urinary problems like kidney failure, septic shock; microcirculatory problems like microangiopathy besides the severe respiratory complications like pneumonia, chest congestion, etc. Along with this, immunogenic activation leading to cytokine storm has a critical effect on weak organs of our body leading to multi-organ failure and even death (Zeng et al. 2020; Connors and Levy 2020; Coperchini et al. 2020; Rodriguez-Morales et al. 2020). There are already many existing amyloidogenic diseases in our body which include both neuropathy and cardiomyopathy and based on the previous findings of amyloidogenicity in SARS-CoV-2 protein and proof of coronavirus proteins accelerating the amyloidogenesis of neurodegenerative protein αS responsible for Parkinson’s Disease (discussed above), it was thought of whether SARS-CoV-2 proteins can affect or aggravate the amyloidogenesis of the other pre-existing amyloidogenic proteins in our body or not (Li et al. 2021). Studies were conducted which yielded many positive results, some of which are discussed in Fig. 3.
Graphical representation of effect of SARS-CoV-2 on pre-existing Amyloidogenic entities in our body; A Fluid-filled alveoli (left) of ARDS affected patient when comes under effect pf COVID-19 results in increased propensity of amyloidosis; B Downregulation of ACE-2 and upregulation of S-protein in lungs post-COVID infection leads to formation of amyloidic microclots in pulmonary vasculature; C Native serum amyloid-A hexamer undergoes several pathways in presence of nine-residue segment of E-protein called SK9 and ultimately leads to amyloid fibril formation; All these results finally culminates into one outcome, i.e., aggravation of COVID-like symptoms and severity of co-morbidities
Amyloidogenesis in COVID-induced ARDS
Acute Respiratory Distress Syndrome (ARDS) is the condition in which fluid gets filled in the air sacs of the lungs, called alveoli, depriving organs of oxygen which is detected in about 20–67% of hospital-admitted COVID patients (Grasselli et al. 2020; Said et al. 2013). Owing to the fact that lung inflammation in many cases can lead to pulmonary as well as non-pulmonary amyloidosis like systemic amyloidosis in pulmonary tuberculosis patients due to SP-C peptide amyloidogenesis. Besides, other conditions like cystic fibrosis, pulmonary sarcoidosis, rheumatic diseases, etc., are prominently related with amyloid A (AA) amyloidosis (Gustafsson et al. 1999; Brunger et al. 2020; Obici and Merlini 2012). All these prior findings led to the assumption of a probable link of amyloidogenesis as the molecular mechanism behind COVID-19 induced ARDS. Though not yet proved, there are many hypothetical pathways that are thought to lead to amyloidogenesis in COVID-induced ARDS, as depicted in Fig. 4. These include over-expression of the enzyme elastase in the plasma which may lead to excessive digestion of elastin proteins in the cell, that may result in formation of amyloidogenic peptides gradually post-COVID infection, overexpression of serum amyloid A (SAA) due to pulmonary inflammation during COVID and subsequent overexpression of matrix metalloprotease enzymes like MMP-3 which can cleave SAA and result in production of amyloidogenic proteins which may cause non-pulmonary amyloidosis, secondary infection post-COVID owing to compromised immunity by pathogens like Klebsiella pneumoniae and Escherichia coli which may lead to the release of lipopolysaccharide-like factors which may induce ARDS due to AA (amyloid-A) amyloidosis, that can severely affect the renal functions in our body (Zahid et al. 2020; Lundmark et al. 2002; Bochicchio et al. 2013). Apart from these reduced redox-homeostasis in lungs post-COVID due to increase in oxidative stress in the pulmonary environment that pertubes the metastable lung surfactant proteins like SP-C resulting in their amyloidogenesis, further signifying the chances of amyloidosis in ARDS patients (Dluhy et al. 2003; Johansson 2001). Due to the above mentioned pathways, the initial amyloids formed may not have such adverse effects in our body but can act as amyloid-enhancing factors which may cause severe amyloidosis in future and result in onset of various amyloidogenic diseases.
The possible pathways of amyloid formation in severe acute respiratory syndrome coronavirus-2/coronavirus disease-2019 (SARS-CoV-2/ COVID-19)-induced acute respiratory distress syndrome (ARDS). Figure taken with permission from Ref. (Sinha and Thakur 2021)
Amyloidogenic microclot formation
Hypercoagulation or microclot formation in the lungs of SARS-CoV-2 infected patients is a common pathology. Since, SARS-CoV-2 achieves host-entry by docking its S-protein with the ACE-2 receptor of the host, the role of both the participants were studied in microclot formation in patients. Angiotensin helps in anti-thrombosis of the platelet, thereby decreasing clot formation in blood, which is catalyzed by ACE-2 enzyme. Due to COVID, downregulation of ACE-2 receptors is noticed which results in microthrombosis of blood in the pulmonary environment (Fraga-Silva et al. 2008; Verdecchia et al. 2020). Furthermore, it was seen that, patients with pre-existing cardiac amyloidosis had an increased rate of microclot formation in their lungs when compared with the ones without cardiac amyloidosis; suggesting that these set of patients are coming under high-risk radar of added morbidities post-COVID infection (Menter et al. 2020; Hanley et al. 2020; Ng et al. 2016). Now, while analyzing the role of S-protein, it must be known beforehand that SARS-CoV-2 can shed the spike protein cover which can circulate to different systems of our body, including the urinary and nervous system, crossing the blood–brain barrier (George et al. 2021; Rhea et al. 2021; Bleu et al. 2015). Healthy platelet-poor plasma (PPP) was tested with and without the addition of S-protein; mass spectrometry, SEM, fluorescence microscopy analysis showed the emergence of dense amyloid structures, resistant to trypsin digestion leading to the formation of microclots, impairing steady blood flow (Grobbelaar et al. 2021; Erickson and Banks 2018). Thus, it was seen that presence of prior cardiac amyloidosis accelerated microclot formation due to the onset of COVID-19 as well as the interaction of S-protein with platelets resulting in amyloid-prone aggregate formation in the pulmonary vasculature post-infection.
Amyloidogenesis in serum amyloid-A protein
Serum amyloid A is such a protein which leads to deposition of amyloid fibrils during the course of many inflammatory diseases like cancer leading to the inflammation, hypercoagulation/thrombosis and also multi-organ damage in many cases. In COVID-19 patients, SAA related amyloidosis like kidney failure and thrombosis is quite common which led to the hypothesis of any direct interaction between SARS-CoV-2 protein and SAA, that may cause long-term risk in COVID survivors in near future like multisystem inflammatory syndrome in children as well as adults (MIS-C and MIS-A, respectively) (Hanff et al. 2020; Fabrizi et al. 2020; Morris et al. 2020; Yeung et al. 2016). Molecular simulation studies proved the effect of 9-residue segment known as SK9 of the E-protein leads to the increased amyloidogenic propensity of SAA. This is guided by mainly three mechanisms: firstly, SK9 binding with whole SAA protein can decrease the stability of native SAA hexamer which results in formation of amyloidogenic monomers; secondly, SK9 can bind with the fragmented SAA and result in formation of amyloid-prone form of SAA; finally, SK9 can stabilize SAA fibrils making them more aggregation prone and resistant to proteolysis (Jana et al. 2021; Zhou et al. 2014; Woo et al. 2007).
Discussion and future prospective
COVID-19, though being primarily a pulmonary viral infection, has several other pathogenesis linked with it like acute respiratory distress syndrome (ARDS) that can result in systemic AA amyloidosis, cytokine storm, heart damage, kidney damage, neurological problems, disturbances in blood flow, etc., (Huang et al. 2021; Lipcsey et al. 2021; Gao et al. 2021; Sinha and Thakur 2021; Sen et al. 2016). Apart from this, hypercoagulation of blood and impaired fibrinolysis have been reported in COVID-19 recovered patients, proposing a link of amyloidogenesis in different proteins of SARS-CoV-2 resulting in amyloidogenic disease-specific symptoms and manifestations in COVID-19 (Grobbelaar et al. 2021). The two most abundant proteins in COVID-19 infected human cells are the N-protein and S-protein of SARS-CoV-2. S-proteins present the primary contact protein between the host and the virus along with being the host-entry point, whereas N-protein is more conserved and stable in the individuals affected than S-protein. Besides, both the proteins have been extensively used as the main antigen for various vaccine productions against SARS-CoV-2.
S-protein amyloidogenesis was tested, and it was proven that endoproteolysis induced by immune-responsive proteases like neutrophil esterase (NE) can nick S-protein at multiple sites and promote amyloid fibril formations; segment 192–212 being the most accurate and pathologically important. Other proteases can also result in such fragmentation followed by amyloidogenesis, but what was proved with NE digestion of S-protein provides the basic mechanism of what might be the case for other enzymes as well. Segment 192–212 in S-protein is that potent segment which is highly amyloidogenic and may result in amyloidosis in the near future after COVID-infection and also after vaccination (Laudicella et al. 2021; de Jong et al. 2006). Next, both S-protein and N-protein were tested against αS protein, amyloidosis of which results in development of Parkinson's Disease (PD), to develop a molecular link between COVID-19 and the onset of Parkinsonism, especially in younger patients post-recovery. S-protein showed no role in enhancement of αS aggregation but N-protein, on the other hand, not only enhanced the aggregation but also resulted in production of more homogenized and stable fibrillar morphology, disturbing the endogenous αS proteastasis in the cell which hampers normal cellular functions and causes Parkinsonism. Therefore, overlapping mechanisms between different pathways of amyloidogenesis in cell and that of ARDS induced cellular machinery, led to the hypothesis of a link between COVID-induced ARDS and amyloidogenesis. Though not molecularly proven, several pathways are present which can lead to amyloidogenesis of different critical proteins like systemic AA, lung surfactant proteins like SP-C, elastin, that may initially cause minute amyloidosis, but in near future can result in production of amyloid-enhancing factors causing severe amyloidogenesis in critical proteins like Transthyretin (TTR) (Thomas et al. 2021; Smith et al. 1979; Koike and Katsuno 2020; Driggin et al. 2020).
At present, worldwide 97 COVID vaccines are in the pipeline, 37 vaccines have been approved/authorized and being used all over the world (Craven 2022; Jeyanathan et al. 2020; Flaxman et al. 2020). Most of the authorized and developing vaccines use either S-protein or N-protein of SARS-CoV-2 as the main antigen, which means apart from the viral infection, we are being medically incorporated with the amyloidogenic viral proteins in the ever-increasing doses of vaccination. Thus, it is of utmost importance to study the side-effects of vaccination using the amyloidogenic viral proteins, as being done in the case of SARS-CoV-2 to prevent the onset of amyloidosis related cardiopathy and neuropathy including neurodegenerative diseases like Alzheimer's disease and Parkinson's disease; seeds of which are being borne in our system from a young age due to COVID-related amyloidogenesis. Summarized data on the amyloidogenic proteins of COVID-19, their mechanism of action and future implications is provided on Table 2.
Conclusion
COVID-19 has been the greatest bane to our existence in recent times and teamed up with the age-old machinery of amyloidogenesis, it is resulting in aggravated complications and morbidities in critical amyloidogenic diseases like ATTR (transthyretin amyloidosis). S-protein and N-protein amyloidogenesis, resulting due to endoproteolysis and proteastasis of cellular αS protein, respectively, may lead to aggregation and amyloid formation that can cause neurodegenerative and other amyloidogenic cardiac and neural complications in future. Along with this, the side effects of COVID-19 vaccination using these very proteins as their main antigen needs to be studied properly to prevent the onset of amyloidogenesis related pathological conditions in individuals post SARS-CoV-2 infection, and aggravation of common morbidities between COVID-19 and critical amyloidogenic diseases like ATTR.
References
Alsaadi EAJ, Neuman BW, Jones IM (2020) Identification of a membrane binding peptide in the envelope protein of MHV Coronavirus. Viruses. https://doi.org/10.3390/v12091054
Bantle CM, Phillips AT, Smeyne RJ, Rocha SM, Olson KE, Tjalkens RB (2019) Infection with mosquito-borne alphavirus induces selective loss of dopaminergic neurons, neuroinflammation and widespread protein aggregation. NPJ Parkinsons Dis 5:20. https://doi.org/10.1038/s41531-019-0090-8
Bar-On YM, Flamholz A, Phillips R, Milo R (2020) Sars-CoV-2 (COVID-19) by the numbers. Elife. https://doi.org/10.7554/eLife.57309
Bhardwaj T, Gadhave K, Kapuganti SK, Kumar P, Brotzakis ZF, Saumya KU, Nayak N, Kumar A, Garg N, Vendruscolo M, Giri R (2021) Amyloidogenic proteins in the SARS-CoV and SARS-CoV-2 proteomes. BioRxiv. https://doi.org/10.1101/2021.05.29.446267
Bleau C, Filliol A, Samson M, Lamontagne L (2015) Brain invasion by mouse hepatitis virus depends on impairment of tight junctions and beta interferon production in brain microvascular endothelial cells. J Virol 89(19):9896–9908. https://doi.org/10.1128/JVI.01501-15
Bochicchio B, Pepe A, Delaunay F, Lorusso M, Baud S, Dauchez M (2013) Amyloidogenesis of proteolytic fragments of human elastin. RSC Adv 3(32):13273–13285. https://doi.org/10.1039/C3RA41893F
Brunger AF, Nienhuis HLA, Bijzet J, Hazenberg BPC (2020) Causes of AA amyloidosis: a systematic review. Amyloid 27(1):1–12. https://doi.org/10.1080/13506129.2019.1693359
Burré J, Sharma M, Südhof TC (2014) α-Synuclein assembles into higher-order multimers upon membrane binding to promote SNARE complex formation. Proc Natl Acad Sci USA 111(40):E4274-4283. https://doi.org/10.1073/pnas.1416598111
Chang MS, Lu YT, Ho ST, Wu CC, Wei TY, Chen CJ, Hsu YT, Chu PC, Chen CH, Chu JM, Jan YL, Hung CC, Fan CC, Yang YC (2004) Antibody detection of SARS-CoV spike and nucleocapsid protein. Biochem Biophys Res Commun 314(4):931–936. https://doi.org/10.1016/j.bbrc.2003.12.195
Charnley M, Islam S, Bindra GK, Engwirda J, Ratcliffe J, Zhou J, Mezzenga R, Hulett MD, Han K, Berryman JT, Reynolds NP (2022) Neurotoxic amyloidogenic peptides in the proteome of SARS-COV2: potential implications for neurological symptoms in COVID-19. Nat Commun 13:3387. https://doi.org/10.1038/s41467-022-30932-1
Chatterjee S, Molenaar R, Tromp L, Wagterveld RM, Roesink HDW, Cornelissen J, Claessens M, Blum C (2021) Optimizing fluorophore density for single virus counting: a photophysical approach. Methods Appl Fluoresc 9(2):025001. https://doi.org/10.1088/2050-6120/abd8e4
Chen SC, Lo SY, Ma HC, Li HC (2009) Expression and membrane integration of SARS-CoV E protein and its interaction with M protein. Virus Genes 38:365–371. https://doi.org/10.1007/s11262-009-0341-6
Clark AH, Judge FJ, Richards JB, Stubbs JM, Sugget A (1981) Electron microscopy of network structures inthermally-induced globular protein gels. Int J Pept Protein Res 17:380–392. https://doi.org/10.1111/j.1399-3011.1981.tb02005.x
Cohen ME, Eichel R, Steiner-Birmanns B, Janah A, Ioshpa M, Bar-Shalom R, Paul JJ, Gaber H, Skrahina V, Bornstein NM, Yahalom G (2020) A case of probable Parkinson’s disease after Sars-CoV-2 infection. Lancet Neurol 19(10):804–805. https://doi.org/10.1016/S1474-4422(20)30305-7
Connors JM, Levy JH (2020) COVID-19 and its implications for thrombosis and anticoagulation. Blood 135(23):2033–2040. https://doi.org/10.1182/blood.2020006000
Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M (2020) The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev 53:25–32. https://doi.org/10.1016/j.cytogfr.2020.05.003
Craven J (2022) COVID-19 vaccine tracker. https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker
Cubuk JAJ, Incicco JJ, Singh S, Stuchell-Brereton MD, Ward MD et al (2021) The Sars-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat Commun. https://doi.org/10.1038/s41467-021-21953-3
de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, do Ha Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J (2006) Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12(10):1203–1207. https://doi.org/10.1038/nm1477
Dimitrov DS, Willey RL, Sato H, Chang LJ, Blumenthal R, Martin MA (1993) Quantitation of humanimmunodeficiency virus type 1 infection kinetics. J Virol 67:2182–2190. https://doi.org/10.1128/JVI.67.4.2182-2190.1993
Dluhy RA, Shanmukh S, Leapard JB, Kruger P, Baatz JE (2003) Deacylated pulmonary surfactant protein SP-C transforms from alpha-helical to amyloid fibril structure via a pH-dependent mechanism: an infrared structural investigation. Biophys J 85(4):2417–2429. https://doi.org/10.1016/s0006-3495(03)74665-7
Dobson CM (1999) Protein misfolding, evolution and disease. Trends Biochem Sci 24(9):329–332. https://doi.org/10.1016/s0968-0004(99)01445-0
Driggin E, Helmke S, De Los SJ, Teruya S, Guadalupe S, Goldsmith J, Maurer MS (2020) Markers of nutritional status and inflammation in transthyretin cardiac amyloidosis: association with outcomes and the clinical phenotype. Amyloid 27(2):73–80. https://doi.org/10.1080/13506129.2019.1698417
Erickson MA, Banks WA (2018) Neuroimmune axes of the blood-brain barriers and blood-brain interfaces: bases for physiological regulation, disease states, and pharmacological interventions. Pharmacol Rev 70(2):278–314. https://doi.org/10.1124/pr.117.014647
Espay AJ, Henderson KK (2011) Postencephalitic parkinsonism and basal ganglia necrosis due to Epstein-Barr virus infection. Neurology 76(17):1529–1530. https://doi.org/10.1212/WNL.0b013e318217e7dd
Fabrizi F, Alfieri CM, Cerutti R, Lunghi G, Messa P (2020) COVID-19 and acute kidney injury: a systematic review and meta-analysis. Pathogens. https://doi.org/10.3390/pathogens9121052
Fakhree MA, Zijlstra N, Raiss CC, Siero CJ, Grabmayr H, Bausch AR, Blum C, Claessens MM (2016) The number of alpha-synuclein proteins per vesicle gives insights into its physiological function. Sci Rep 6:30658. https://doi.org/10.1038/srep30658
Fakhree MAA, Nolten IS, Blum C, Claessens M (2018) Different conformational subensembles of the intrinsically disordered protein alpha-synuclein in cells. J Phys Chem Lett 9(6):1249–1253. https://doi.org/10.1021/acs.jpclett.8b00092
Fishman PS, Gass JS, Swoveland PT, Lavi E, Highkin MK, Weiss SR (1985) Infection of the basal ganglia by a murine coronavirus. Science 229(4716):877–879. https://doi.org/10.1126/science.2992088
Flaxman S, Mishra S, Gandy A, Unwin HJT, Mellan TA, Coupland H, Whittaker C, Zhu H, Berah T, Eaton JW, Monod M, Imperial College COVID-19 Response Team, Ghani AC, Donnelly CA, Riley S, Vollmer MAC, Ferguson NM, Okell LC, Bhatt S (2020) Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature 584(7820):257–261. https://doi.org/10.1038/s41586-020-2405-7
Fraga-Silva RA, Pinheiro SV, Goncalves AC, Alenina N, Bader M, Santos RA (2008) The antithrombotic effect of angiotensin-(1–7) involves mas-mediated NO release from platelets. Mol Med 14(1–2):28–35. https://doi.org/10.2119/2007-00073.Fraga-Silva
Galkin AP (2021) Hypothesis: AA amyloidosis is a factor causing systemic complications after coronavirus disease. Prion 15(1):53–55. https://doi.org/10.1080/19336896.2021.1910468
Galzitskaya OV, Garbuzynskiy SO, Lobanov MY (2006) Prediction of amyloidogenic and disordered regions in protein chains. PLOS Comput Biol 2:e177. https://doi.org/10.1371/journal.pcbi.0020177
Gao YM, Xu G, Wang B, Liu BC (2021) Cytokine storm syndrome in coronavirus disease 2019: a narrative review. J Intern Med 289(2):147–161. https://doi.org/10.1111/joim.13144
George S, Pal AC, Gagnon J, Timalsina S, Singh P, Vydyam P, Munshi M, Chiu JE, Renard I, Harden CA, Ott IM, Watkins AE, Vogels CBF, Lu P, Tokuyama M, Venkataraman A, Casanovas-Massana A, Wyllie AL, Rao V, Campbell M, Farhadian SF, Grubaugh ND, Dela Cruz CS, Ko AI, Berna Perez AZ, Akaho EH, Moledina DG, Testani J, John AR, Ledizet M, Mamoun CB, the Yale IT, (2021) Evidence for Sars-CoV-2 spike protein in the urine of COVID-19 patients. Kidney 2(6):924–936. https://doi.org/10.34067/KID.0002172021
Ghetti B, Piccardo P, Frangione B, Bugiani O, Giaccone G, Young K, Prelli F, Farlow MR, Dlouhy SR, Tagliavini F (1996) Prion protein amyloidosis. Brain Pathol 6(2):127–145. https://doi.org/10.1111/j.1750-3639.1996.tb00796.x
Ghiso J, Frangione B (2002) Amyloidosis and Alzheimer’s disease. Adv Drug Deliv Rev 54(12):1539–1551. https://doi.org/10.1016/s0169-409x(02)00149-7
Ghosh A, Pithadia AS, Bhat J, Bera S, Midya A, Fierke CA, Ramamoorthy A, Bhunia A (2015) Self-assembly of a nine-residue amyloid-forming peptide fragment of SARS corona virus E-protein: mechanism of self aggregation and amyloid-inhibition of hIAPP. Biochemistry 54(13):2249–2261. https://doi.org/10.1021/acs.biochem.5b00061
Grasselli G, Tonetti T, Protti A, Langer T, Girardis M, Bellani G, Laffey J, Carrafiello G, Carsana L, Rizzuto C, Zanella A, Scaravilli V, Pizzilli G, Grieco DL, Di Meglio L, de Pascale G, Lanza E, Monteduro F, Zompatori M, Filippini C, Locatelli F, Cecconi M, Fumagalli R, Nava S, Vincent JL, Antonelli M, Slutsky AS, Pesenti A, Ranieri VM, collaborators, (2020) Pathophysiology of COVID-19-associated acute respiratory distress syndrome: a multicentre prospective observational study. Lancet Respir Med 8(12):1201–1208. https://doi.org/10.1016/S2213-2600(20)30370-2
Grobbelaar LM, Venter C, Vlok M, Ngoepe M, Laubscher GJ, Lourens PJ, Steenkamp J, Kell DB, Pretorius E (2021) Sars-CoV-2 spike protein S1 induces fibrin(ogen) resistant to fibrinolysis: implications for microclot formation in COVID-19. Biosci Rep. https://doi.org/10.1042/BSR20210611
Gustafsson M, Thyberg J, Naslund J, Eliasson E, Johansson J (1999) Amyloid fibril formation by pulmonary surfactant protein C. FEBS Lett 464(3):138–142. https://doi.org/10.1016/s0014-5793(99)01692-0
Halverson K, Fraser PE, Kirschner DA, Lansbury PT Jr (1990) Molecular determinants of amyloid deposition in Alzheimer’s disease: conformational studies of synthetic beta-protein fragments. Biochemistry 29(11):2639–2644. https://doi.org/10.1021/bi00463a003
Hamodrakas SJ, Liappa C, Iconomidou VA (2007) Consensus prediction of amyloidogenic determinantsin amyloid fibril-forming proteins. Int J Biol Macromol 41:295–300. https://doi.org/10.1016/j.ijbiomac.2007.03.008
Hanff TC, Mohareb AM, Giri J, Cohen JB, Chirinos JA (2020) Thrombosis in COVID-19. Am J Hematol 95(12):1578–1589. https://doi.org/10.1002/ajh.25982
Hanley B, Lucas SB, Youd E, Swift B, Osborn M (2020) Autopsy in suspected COVID-19 cases. J Clin Pathol 73(5):239–242. https://doi.org/10.1136/jclinpath-2020-206522
Huang C, Huang L, Wang Y, Li X, Ren L, Gu X, Kang L, Guo L, Liu M, Zhou X, Luo J, Huang Z, Tu S, Zhao Y, Chen L, Xu D, Li Y, Li C, Peng L, Li Y, Xie W, Cui D, Shang L, Fan G, Xu J, Wang G, Wang Y, Zhong J, Wang C, Wang J, Zhang D, Cao B (2021) 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet 397(10270):220–232. https://doi.org/10.1016/S0140-6736(20)32656-8
Jana AK, Greenwood AB, Hansmann UHE (2021) Presence of a SARS-COV-2 protein enhances amyloid formation of serum amyloid A. BioRxiv. https://doi.org/10.1101/2021.05.18.444723
Jeyanathan M, Afkhami S, Smaill F (2020) Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol 20:615–632. https://doi.org/10.1038/s41577-020-00434-6
Johansson J (2001) Membrane properties and amyloid fibril formation of lung surfactant protein C. Biochem Soc Trans 29(Pt 4):601–606. https://doi.org/10.1042/bst0290601
Johansson C, Kirsebom FCM (2021) Neutrophils in respiratory viral infections. Mucosal Immunol 14(4):815–827. https://doi.org/10.1038/s41385-021-00397-4
Kaur U, Lee JC (2020) Unroofing site-specific alpha-synuclein-lipid interactions at the plasma membrane. Proc Natl Acad Sci USA 117(32):18977–18983. https://doi.org/10.1073/pnas.2006291117
Koike H, Katsuno M (2020) Transthyretin amyloidosis: update on the clinical spectrum, pathogenesis, and disease-modifying therapies. Neurol Ther 9(2):317–333. https://doi.org/10.1007/s40120-020-00210-7
Laudicella R, Burger IA, Panasiti F, Longo C, Scalisi S, Minutoli F, Baldari S, Grimaldi LME, Alongi P (2021) Subcutaneous uptake on [18F]Florbetaben PET/CT: a case report of possible amyloid-beta immune-reactivity after COVID-19 vaccination. SN Compr Clin Med 3(12):2626–2628. https://doi.org/10.1007/s42399-021-01058-0
Lautenschläger J, Kaminski CF, Kaminski Schierle GS (2017) α-Synuclein - regulator of exocytosis, endocytosis, or both? Trends Cell Biol 27(7):468–479. https://doi.org/10.1016/j.tcb.2017.02.002
Lawrence DS, Jiang T, Levett M (1995) Self-assembling supramolecular complexes. Chem Rev 95:2229–2260. https://doi.org/10.1021/cr00038a018
Lee YN, Chen LK, Ma HC, Yang HH, Li HP, Lo SY (2005) Thermal aggregation of SARS-CoV membrane protein. J Virol Methods 129(2):152–161. https://doi.org/10.1016/j.jviromet.2005.05.022
Lee JG, Huang W, Lee H, van de Leemput J, Kane MA, Han Z (2021) Characterization of SARS-CoV-2 proteins reveals Orf6 pathogenicity, subcellular localization, host interactions and attenuation by Selinexor. Cell Biosci 11(1):58. https://doi.org/10.1186/s13578-021-00568-7
Li Y, Surya W, Claudine S, Torres J (2014) Structure of a conserved Golgi complex-targeting signal in coronavirus envelope proteins. J Biol Chem 289(18):12535–12549. https://doi.org/10.1074/jbc.M114.560094
Li J, Huang DQ, Zou B, Yang H, Hui WZ, Rui F, Yee NTS, Liu C, Nerurkar SN, Kai JCY, Teng MLP, Li X, Zeng H, Borghi JA, Henry L, Cheung R, Nguyen MH (2021) Epidemiology of COVID-19: a systematic review and meta-analysis of clinical characteristics, risk factors, and outcomes. J Med Virol 93(3):1449–1458. https://doi.org/10.1002/jmv.26424
Lipcsey M, Persson B, Eriksson O, Blom AM, Fromell K, Hultstrom M, Huber-Lang M, Ekdahl KN, Frithiof R, Nilsson B (2021) The outcome of critically ill COVID-19 patients is linked to thromboinflammation dominated by the kallikrein/kinin system. Front Immunol 12:627579. https://doi.org/10.3389/fimmu.2021.627579
Lopez de la Paz M, Serrano L (2004) Sequence determinants of amyloid fibril formation. Proc Natl Acad Sci USA 101(1):87–92. https://doi.org/10.1073/pnas.2634884100
Lu K, Jacob J, Thiyagarajan P, Conticello VP, Lynn DG (2003) Exploiting amyloid fibril lamination for nanotube self-assembly. J Am Chem Soc 125(21):6391–6393. https://doi.org/10.1021/ja0341642
Lundmark K, Westermark GT, Nystrom S, Murphy CL, Solomon A, Westermark P (2002) Transmissibility of systemic amyloidosis by a prion-like mechanism. Proc Natl Acad Sci USA 99(10):6979–6984. https://doi.org/10.1073/pnas.092205999
Marzban L, Park K, Verchere CB (2003) Islet amyloid polypeptide and type 2 diabetes. Exp Gerontol 38(4):347–351. https://doi.org/10.1016/s0531-5565(03)00004-4
Maurer-Stroh S, Debulpaep M, Kuemmerer N, Lopez de la Paz M, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L, Schymkowitz JW, Rousseau F (2010) Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods 7(3):237–242. https://doi.org/10.1038/nmeth.1432
Menter T, Haslbauer JD, Nienhold R, Savic S, Hopfer H, Deigendesch N, Frank S, Turek D, Willi N, Pargger H, Bassetti S, Leuppi JD, Cathomas G, Tolnay M, Mertz KD, Tzankov A (2020) Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology 77(2):198–209. https://doi.org/10.1111/his.14134
Merello M, Bhatia KP, Obeso JA (2021) Sars-CoV-2 and the risk of Parkinson’s disease: facts and fantasy. Lancet Neurol 20(2):94–95. https://doi.org/10.1016/S1474-4422(20)30442-7
Michiels E, Rousseau F, Schymkowitz J (2021) Mechanisms and therapeutic potential of interactions between human amyloids and viruses. Cell Mol Life Sci 78(6):2485–2501. https://doi.org/10.1007/s00018-020-03711-8
Minor D, Kim P (1994) Measurement of the β-sheet-forming propensities of amino acids. Nature 367:660–663. https://doi.org/10.1038/367660a0
Morris SB, Schwartz NG, Patel P, Abbo L, Beauchamps L, Balan S, Lee EH, Paneth-Pollak R, Geevarughese A, Lash MK, Dorsinville MS, Ballen V, Eiras DP, Newton-Cheh C, Smith E, Robinson S, Stogsdill P, Lim S, Fox SE, Richardson G, Hand J, Oliver NT, Kofman A, Bryant B, Ende Z, Datta D, Belay E, Godfred-Cato S (2020) Case series of multisystem inflammatory syndrome in adults associated with Sars-CoV-2 infection - United Kingdom and United States, March-August 2020. MMWR Morb Mortal Wkly Rep 69(40):1450–1456. https://doi.org/10.15585/mmwr.mm6940e1
Munch J, Rucker E, Standker L, Adermann K, Goffinet C, Schindler M, Wildum S, Chinnadurai R, Rajan D, Specht A, Gimenez-Gallego G, Sanchez PC, Fowler DM, Koulov A, Kelly JW, Mothes W, Grivel JC, Margolis L, Keppler OT, Forssmann WG, Kirchhoff F (2007) Semen-derived amyloid fibrils drastically enhance HIV infection. Cell 131(6):1059–1071. https://doi.org/10.1016/j.cell.2007.10.014
Nemani VM, Lu W, Berge V, Nakamura K, Onoa B, Lee MK, Chaudhry FA, Nicoll RA, Edwards RH (2010) Increased expression of alpha-Synuclein reduces neurotransmitter release by inhibiting synapticvesicle reclustering after endocytosis. Neuro 65:66–79. https://doi.org/10.1016/j.neuron.2009.12.023
Ng DL, Al Hosani F, Keating MK, Gerber SI, Jones TL, Metcalfe MG, Tong S, Tao Y, Alami NN, Haynes LM, Mutei MA, Abdel-Wareth L, Uyeki TM, Swerdlow DL, Barakat M, Zaki SR (2016) Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of middle east respiratory syndrome coronavirus infection in the United Arab Emirates. Am J Pathol 186(3):652–658. https://doi.org/10.1016/j.ajpath.2015.10.024
Nyström S, Hammarström P (2021) Amyloidogenesis of Sars-CoV-2 spike protein. BioRxiv. https://doi.org/10.1101/2021.12.16.472920
Obici L, Merlini G (2012) AA amyloidosis: basic knowledge, unmet needs and future treatments. Swiss Med Wkly 142:13580. https://doi.org/10.4414/smw.2012.13580
Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, Kugathasan R, Penn R, Brown JC, Sanchez-David RY, Braga L, Williamson MK, Hassard JA, Staller E, Hanley B, Osborn M, Giacca M, Davidson AD, Matthews DA, Barclay WS (2021) The furin cleavage site in the Sars-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol 6(7):899–909. https://doi.org/10.1038/s41564-021-00908-w
Perozzo R, Folkers G, Scapozza L (2004) Thermodynamics of protein-ligand interactions: history, presence, and future aspects. J Recept Signal Transduct Res 24(1–2):1–52. https://doi.org/10.1081/rrs-120037896
Petrou M, Dwamena BA, Foerster BR, MacEachern MP, Bohnen NI, Muller ML, Albin RL, Frey KA (2015) Amyloid deposition in Parkinson’s disease and cognitive impairment: a systematic review. Mov Disord 30(7):928–935. https://doi.org/10.1002/mds.26191
Rangan R, Zheludev IN, Hagey RJ, Pham EA, Wayment-Steele HK, Glenn JS, Das R (2020) RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA 26(8):937–959. https://doi.org/10.1261/rna.076141.120
Rhea EM, Logsdon AF, Hansen KM, Williams LM, Reed MJ, Baumann KK, Holden SJ, Raber J, Banks WA, Erickson MA (2021) The S1 protein of Sars-CoV-2 crosses the blood-brain barrier in mice. Nat Neurosci 24(3):368–378. https://doi.org/10.1038/s41593-020-00771-8
Rodriguez-Morales AJ, Cardona-Ospina JA, Gutiérrez-Ocampo E, Villamizar-Peña R, Holguin-Rivera Y, Escalera-Antezana JP, Alvarado-Arnez LE, Bonilla-Aldana DK, Franco-Paredes C, Henao-Martinez AF, Paniz-Mondolfi A, Lagos-Grisales GJ, Ramírez-Vallejo E, Suárez JA, Zambrano LI, Villamil-Gómez WE, Balbin-Ramon GJ, Rabaan AA, Harapan H, Dhama K, Nishiura H, Kataoka H, Ahmad T, Sah R (2020) Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis 34:101623. https://doi.org/10.1016/j.tmaid.2020.101623
Said SM, Sethi S, Valeri AM, Leung N, Cornell LD, Fidler ME, Herrera Hernandez L, Vrana JA, Theis JD, Quint PS, Dogan A, Nasr SH (2013) Renal amyloidosis: origin and clinicopathologic correlations of 474 recent cases. Clin J Am Soc Nephrol 8(9):1515–1523. https://doi.org/10.2215/CJN.10491012
Sarkar N (2020) Chapter 12 - functional amyloids. In: Pal K, Banerjee I, Sarkar P et al (eds) Biopolymer-based formulations. Elsevier, Amsterdam, pp 263–282. https://doi.org/10.1016/B978-0-12-816897-4.00012-6
Semerdzhiev SA, Fakhree MAA, Segers-Nolten I, Blum C, Claessens MMAE (2022) Interactions between Sars-CoV-2 N-Protein and α-Synuclein Accelerate Amyloid Formation. ACS Chem Neurosci 13(1):143–150. https://doi.org/10.1021/acschemneuro.1c00666
Sen ES, Clarke SL, Ramanan AV (2016) Macrophage activation syndrome. Indian J Pediatr 83(3):248–253. https://doi.org/10.1007/s12098-015-1877-1
Shu-Qun Liu X-LJ, Tao Y, Tan D-Y, Zhang K-Q, Yun-Xin Fu (2012) Protein folding, binding and energy landscape: a synthesis. Protein Engineering. InTech, London. https://doi.org/10.5772/30440
Sinha N, Thakur AK (2021) Likelihood of amyloid formation in COVID-19-induced ARDS. Trends Microbiol 29(11):967–969. https://doi.org/10.1016/j.tim.2021.03.008
Sipe DJ (2008) Amyloid proteins: the beta sheet conformation and disease. Wiley 1:799
Smith RR, Hutchins GM, Moore GW, Humphrey RL (1979) Type and distribution of pulmonary parenchymal and vascular amyloid. Correlation with cardiac amyloid. Am J Med 66(1):96–104. https://doi.org/10.1016/0002-9343(79)90488-1
Strnad P, McElvaney NG, Lomas DA (2020) Alpha1-antitrypsin deficiency. N Engl J Med 382(15):1443–1455. https://doi.org/10.1056/NEJMra1910234
Taquet M, Geddes JR, Husain M, Luciano S, Harrison PJ (2021) 6-month neurological and psychiatric outcomes in 236379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatry 8:416–427. https://doi.org/10.1016/s2215-0366(21)00084-5
Tayeb-Fligelman E, Cheng X, Tai C, Bowler JT, Griner S, Sawaya MR, Seidler PM, Jiang YX, Lu J, Rosenberg GM, Salwinski L, Abskharon R, Zee CT, Hou K, Li Y, Boyer DR, Murray KA, Falcon G, Anderson DH, Cascio D, Saelices L, Damoiseaux R, Guo F, Eisenberg DS (2021) Inhibition of amyloid formation of the nucleoprotein of Sars-CoV-2. BioRxiv. https://doi.org/10.1101/2021.03.05.434000
Thomas HMA-G, Berk JL, Chiara B, Vera B, Teresa C, Thibaud D, Angela D, Brian MD, Nowell F, Hanna KG, Morie G, Julian DG, Esther G, Mazen H, David RH, Sami LK, Mathew SM, Jose N-N, Kemi O, Luis FQ, Andrew MR, Hartmut HS, Jacqueline S, Marcia W-C, Carol W, Frederick LR (2021) ATTR amyloidosis during the COVID-19 pandemic: insights from a global medical roundtable. Orphanet J Rare Dis. https://doi.org/10.1186/s13023-021-01834-0
Timani KA, Liao Q, Ye L, Zeng Y, Liu J, Zheng Y, Ye L, Yang X, Lingbao K, Gao J, Zhu Y (2005) Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res 114(1–2):23–34. https://doi.org/10.1016/j.virusres.2005.05.007
Upadhyay V, Lucas A, Panja S, Miyauchi R, Mallela KMG (2021) Receptor binding, immune escape, and protein stability direct the natural selection of Sars-CoV-2 variants. J Biol Chem 297(4):101208. https://doi.org/10.1016/j.jbc.2021.101208
Verdecchia P, Cavallini C, Spanevello A, Angeli F (2020) The pivotal link between ACE2 deficiency and Sars-CoV-2 infection. Eur J Intern Med 76:14–20. https://doi.org/10.1016/j.ejim.2020.04.037
Woo PC, Wang M, Lau SK, Xu H, Poon RW, Guo R, Wong BH, Gao K, Tsoi HW, Huang Y, Li KS, Lam CS, Chan KH, Zheng BJ, Yuen KY (2007) Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol 81(4):1574–1585. https://doi.org/10.1128/JVI.02182-06
Yeung ML, Yao Y, Jia L, Chan JF, Chan KH, Cheung KF, Chen H, Poon VK, Tsang AK, To KK, Yiu MK, Teng JL, Chu H, Zhou J, Zhang Q, Deng W, Lau SK, Lau JY, Woo PC, Chan TM, Yung S, Zheng BJ, Jin DY, Mathieson PW, Qin C, Yuen KY (2016) MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat Microbiol 1(3):16004. https://doi.org/10.1038/nmicrobiol.2016.4
Zahid U, Ramachandran P, Spitalewitz S, Alasadi L, Chakraborti A, Azhar M, Mikhalina G, Sherazi A, Narh JT, Khattar P, Bedi P (2020) Acute Kidney Injury in COVID-19 Patients: An Inner City Hospital Experience and Policy Implications. Am J Nephrol 51(10):786–796. https://doi.org/10.1159/000511160
Zeng JH, Wu WB, Qu JX, Wang Y, Dong CF, Luo YF, Zhou D, Feng WX, Feng C (2020) Cardiac manifestations of COVID-19 in Shenzhen. China Infection 48(6):861–870. https://doi.org/10.1007/s15010-020-01473-w
Zhang SM, Liao Y, Neo TL, Lu Y, Liu DX, Vahlne A, Tam JP (2018) Identification and application of self-binding zipper-like sequences in SARS-CoV spike protein. Int J Biochem Cell Biol 101:103–112. https://doi.org/10.1016/j.biocel.2018.05.012
Zhou J, Chu H, Li C, Wong BH, Cheng ZS, Poon VK, Sun T, Lau CC, Wong KK, Chan JY, Chan JF, To KK, Chan KH, Zheng BJ, Yuen KY (2014) Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis 209(9):1331–1342. https://doi.org/10.1093/infdis/jit504
Acknowledgements
The authors acknowledge the infrastructural facilities provided by NIT Rourkela.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Seth, P., Sarkar, N. A comprehensive mini-review on amyloidogenesis of different SARS-CoV-2 proteins and its effect on amyloid formation in various host proteins. 3 Biotech 12, 322 (2022). https://doi.org/10.1007/s13205-022-03390-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03390-1