Abstract.
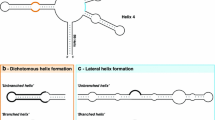
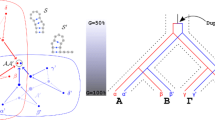
We suggest a nucleotide substitution model that takes correlation between base-paired nucleotides into account. The model includes the estimation of the transition–transversion ratio and allows inference of the shape parameter of a discrete gamma distribution to include rate heterogeneity. A Cox-test statistic, applied to a diatom ribosomal RNA alignment, shows that the suggested correlation model explains evolution of the stem region better than usual independence models. Moreover, the Cox-test procedure is extended to shed some light upon the problem of assigning helical regions in a secondary structure based alignment. This approach provides an estimate of the percentage of stem positions that do not appear to be correlated.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 4 March 1999 / Accepted: 10 May 1999
Rights and permissions
About this article
Cite this article
Schöniger, M., von Haeseler, A. Toward Assigning Helical Regions in Alignments of Ribosomal RNA and Testing the Appropriateness of Evolutionary Models. J Mol Evol 49, 691–698 (1999). https://doi.org/10.1007/PL00006590
Issue Date:
DOI: https://doi.org/10.1007/PL00006590