Abstract
KIF1Bβ is a kinesin-like, microtubule-based molecular motor protein involved in anterograde axonal vesicular transport in vertebrate and invertebrate neurons. Certain KIF1Bβ isoforms have been implicated in different forms of human neurodegenerative disease, with characterization of their functional integration and regulation in the context of synaptic signaling still ongoing. Here, we characterize human KIF1Bβ (isoform NM015074), whose expression we show to be developmentally regulated and elevated in cortical areas of the CNS (including the motor cortex), in the hippocampus, and in spinal motor neurons. KIF1Bβ localizes to the cell body, axon, and dendrites, overlapping with synaptic-vesicle and postsynaptic-density structures. Correspondingly, in purified cortical synaptoneurosomes, KIF1Bβ is enriched in both pre- and postsynaptic structures, forming detergent-resistant complexes. Interestingly, KIF1Bβ forms RNA–protein complexes, containing the dendritically localized Arc and Calmodulin mRNAs, proteins previously shown to be part of RNA transport granules such as Purα, FMRP and FXR2P, and motor protein KIF3A, as well as Calmodulin. The interaction between KIF1Bβ and Calmodulin is Ca+2-dependent and takes place through a domain mapped at the carboxy-terminal tail of the motor. Live imaging of cortical neurons reveals active movement by KIF1Bβ at dendritic processes, suggesting that it mediates the transport of dendritically localized mRNAs. Finally, we show that synaptic recruitment of KIF1Bβ is activity-dependent and increased by stimulation of metabotropic or ionotropic glutamate receptors. The activity-dependent synaptic recruitment of KIF1Bβ, its interaction with Ca2+ sensor Calmodulin, and its new role as a dendritic motor of ribonucleoprotein complexes provide a novel basis for understanding the concerted co-ordination of motor protein mobilization and synaptic signaling pathways.









Similar content being viewed by others
Abbreviations
- Arc:
-
Activity-regulated cytoskeleton-associated protein
- CaM:
-
Calmodulin
- CMT2A:
-
Charcot-Marie-Tooth type 2A motor neuron disease
- DHPG:
-
S-3,5,-dihydroxyphenylglycine
- DMEM:
-
Dulbecco’s Modified Eagle’s Medium
- FMRP:
-
Fragile X mental retardation protein
- FXR2P:
-
Fragile X-related protein 2
- HBSS:
-
Hank’s balanced salt solution
- KIF1Bβ:
-
Kinesin family member 1Bβ
- MEM:
-
Minimal essential medium (Eagle’s)
- Purα:
-
Purine-rich element binding protein alpha
- RT-PCR:
-
Reverse-transcription polymerase chain reaction
References
Lawrence CJ et al (2004) A standardized kinesin nomenclature. J Cell Biol 167:19–22
Nangaku M et al (1994) KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell 79:1209–1220
Mok H, Shin H, Kim S, Lee JR, Yoon J, Kim E (2002) Association of the kinesin superfamily motor protein KIF1Balpha with postsynaptic density-95 (PSD-95), synapse-associated protein-97, and synaptic scaffolding molecule PSD-95/discs large/zona occludens-1 proteins. J Neurosci 22:5253–5258
Zhao C et al (2001) Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell 105:587–597
Zuchner S et al (2004) Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet 36:449–451
Pantelidou M, Zographos SE, Lederer CW, Kyriakides T, Pfaffl MW, Santama N (2007) Differential expression of molecular motors in the motor cortex of sporadic ALS. Neurobiol Dis 26:577–589
Hall DH, Hedgecock EM (1991) Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65:837–847
Klopfenstein DR, Vale RD (2004) The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans. Mol Biol Cell 15:3729–3739
Aulchenko YS et al (2008) Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet 40:1402–1403
Booth DR et al (2010) Lack of support for association between the KIF1B rs10492972[C] variant and multiple sclerosis. Nat Genet 42:469–470
Hintzen RQ, AulchenkoYS, Ramagopalan S, Ebers, GC, van Duijn CM (2010) Reply to “Lack of support for association between the KIF1B rs10492972[C] variant and multiple sclerosis”. Nat Genet 40:1402–1403
Lyons DA, Naylor SG, Scholze A, Talbot WS (2009) Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat Genet 41:854–858
Schlisio S et al (2008) The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev 22:884–893
Yeh IT et al (2008) A germline mutation of the KIF1B beta gene on 1p36 in a family with neural and nonneural tumors. Hum Genet 124:279–285
Munirajan AK et al (2008) KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J Biol Chem 283:24426–24434
Zhang H et al (2010) Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet 42:755–758
Ferrari F, Mercaldo V, Piccoli G, Sala C, Cannata S, Achsel T, Bagni C (2007) The fragile X mental retardation protein-RNP granules show an mGluR1-dependent localization in the post-synaptic spines. Mol Cell Neurosci 34:343–354
Dotti CG, Sullivan CA, Banker GA (1988) The establishment of polarity by hippocampal neurons in culture. J Neurosci 8:1454–1468
Santama N, Krijnse-Locker J, Griffiths G, Noda Y, Hirokawa N, Dotti CG (1998) KIF2beta, a new kinesin superfamily protein in non-neuronal cells, is associated with lysosomes and may be implicated in their centrifugal translocation. EMBO J 17:5855–5867
Nagy A, Delgado-Escueta AV (1984) Rapid preparation of synaptosomes from mammalian brain using nontoxic isoosmotic gradient material (Percoll). J Neurochem 43:1114–1123
Pilo BP et al (2007) Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J 26:2991–3002
Napoli I et al (2008) The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134:1042–1054
Phillips GR et al (2001) The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron 32:63–77
Di Penta. A et al (2009) Dendritic LSm1/CBP80-mRNPs mark the early steps of transport commitment and translational control. J Cell Biol 184:423–435
Christodoulou A, Lederer CW, Surrey T, Vernos I, Santama N (2006) Motor protein KIFC5A interacts with Nubp1 and Nubp2, and is implicated in the regulation of centrosome duplication. J Cell Sci 119:2035–2047
Bolte S, Cordelieres FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232
Sans N, Wang PY, Du Q, Petralia RS, Wang YX, Nakka S, Blumer JB, Macara IG, Wenthold RJ (2005) mPins modulates PSD-95 and SAP102 trafficking and influences NMDA receptor surphase expression. Nat Cell Biol 7:1179–1190
Rashbad WS (2007-2009) ImageJ. U.S National Institutes of Health, Bethesda
Kanai Y, Dohmae N, Hirokawa N (2004) Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43:513–525
Elvira G et al (2006) Characterization of an RNA granule from developing brain. Mol Cell Proteom 5:635–651
Cashman NR et al (1992) Neuroblastoma × spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn 194:209–221
Titulaer MN, Ghijsen WE (1997) Synaptoneurosomes. A preparation for studying subhippocampal GABAA receptor activity. Methods Mol Biol 72:49–59
Berchtold MW, Egli R, Rhyner JA, Hameister H, Strehler EE (1993) Localization of the human bona fide calmodulin genes CALM1, CALM2, and CALM3 to chromosomes 14q24-q31, 2p21.1-p21.3, and 19q13.2-q13.3. Genomics 16:461–465
Friedberg F, Rhoads AR (2001) Sequence homology of the 3′-untranslated region of calmodulin III in mammals. Mol Biol Rep 28:27–30
Falley K et al (2009) Shank1 mRNA: dendritic transport by kinesin and translational control by the 5′ untranslated region. Traffic 10:844–857
Hirokawa N (2006) mRNA transport in dendrites: RNA granules, motors, and tracks. J Neurosci 26:7139–7142
Martin KC, Zukin RS (2006) RNA trafficking and local protein synthesis in dendrites: an overview. J Neurosci 26:7131–7134
Gallia GL, Darbibian N, Johnson EM, Khalili K (2000) Self-association of Purα is mediated by RNA. J Cell Biochem 74:334–348
Ohashi S, Koike K, Omori A, Ichinose S, Ohara S, Kobayashi S, Sato TA, Anzai K (2002) Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin VA and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem 277:37804–37810
Brown V, Small K, Lakkis L, Feng Y, Gunter C, Wilkinson KD, Warren ST (1998) Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J Biol Chem 273:15521–15527
Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ (2005) Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav 4:350–359
Ceman S, Brown V, Warren ST (1999) Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol Cell Biol 19:7925–7932
Link W et al (1995) Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci USA 92:5734–5738
Lyford GL et al (1995) Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 14:433–445
Steward O, Wallace CS, Lyford GL, Worley PF (1998) Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21:741–751
Vehey KJ, Hammond JW (2009) Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol 10:765–777
Ma B, Savas JN, Yu MS, Culver BP, Chao MV, Tanese N (2011) Huntingtin mediates dendritic transport of β-actin mRNA in rat neurons. Sci Rep 1:140
Guillaud L, Setou M, Hirokawa N (2003) KIF17 dynamics and regulation of NR2B trafficking in hippocampal neurons. J Neurosci 23:131–140
Rook MS, Lu M, Kosik KS (2000) CaMKII 3 untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci 20:6385–6393
Savas JN, Ma B, Deinhardt K, Culver BP, Restituito S, Wu L, Belasco JG, Chao MV, Tanese N (2010) A role for huntington disease protein in dendritic RNA granules. J Biol Chem 285:13142–13153
Hirokawa N, Niwa S, Tanaka Y (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68:610–638
Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N (1995) The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 81:769–780
Hammond JW, Cai D, Blasius LT, Li Z, Jiang Y, Jih GT, Meyhofer E, Verhey KJ (2009) Mammalian kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. Plos Biol 7:e72
Niwa S, Tanaka Y, Hirokawa N (2008) KIF1Bbeta- and KIF1A-mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD. Nat Cell Biol 10:1269–1279
Kumar J, Choudhary BC, Metpally R, Zheng Q, Nonet ML, Ramanathan S, Klopferstein DR, Koushika SP (2010) The Caenorhadditis elegans kinesin-3 motor UNC-104/KIF1A is degraded upon loss of specific binding to cargo. Plos Genetics 6:1–17
Wagner OI, Esposito A, Köhler B, Chen C-W, Shen C-P, Wu G-H, Butkevich E, Mandalapu S, Wenzel D, Wouters FS, Klopfenstein DR (2009) Synaptic scaffolding protein SYD-2 clusters and activates kinesin-3 UNC-104 in C. elegans. Proc Natl Acad Sci USA 106:19605–19610
Xue X, Jaulin F, Espenel C, Kreitzer G (2010) PH-domain-dependent selective transport of p75 by kinesin-3 family motors in non-polarized MDCK cells. J Cell Sci 123:1732–1741
Cai Q, Sheng ZH (2009) Molecular motors and synaptic assembly. Neuroscientist 15:78–89
Matthies HJ, Miller RJ, Palfrey HC (1993) Calmodulin binding to and cAMP-dependent phosphorylation of kinesin light chains modulate kinesin ATPase activity. J Biol Chem 268:11176–11187
Nakamura N, Miyake Y, Matsushita M, Tanaka S, Inoue H, Kanazawa H (2002) KIF1Bbeta2, capable of interacting with CHP, is localized to synaptic vesicles. J Biochem 132:483–491
Rusnak F, Mertz P (2000) Calcineurin: form and function. Physiol Rev 80:1483–1521
Macaskill AF et al (2009) Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron 61:541–555
Wang X, Schwarz TL (2009) The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell 136:163–174
Reddy VS, Reddy AS (2002) The calmodulin-binding domain from a plant kinesin functions as a modular domain in conferring Ca2+-calmodulin regulation to animal plus- and minus-end kinesins. J Biol Chem 277:48058–48065
Moga DE, Calhoun ME, Chowdhury A, Worley P, Morrison JH, Shapiro ML (2004) Activity-regulated cytoskeletal-associated protein is localized to recently activated excitatory synapses. Neuroscience 125:7–11
Rodriguez JJ et al (2005) Long-term potentiation in the rat dentate gyrus is associated with enhanced Arc/Arg3.1 protein expression in spines, dendrites and glia. Eur J Neurosci 21:2384–2396
Bramham CR, Worley PF, Moore MJ, Guzowski JF (2008) The immediate early gene arc/arg3.1: regulation, mechanisms, and function. J Neurosci 28:11760–11767
Chowdhury S et al (2006) Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron 52:445–459
Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT (2006) Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron 52:461–474
Shepherd JD et al (2006) Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52:475–484
Messaoudi E et al (2007) Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci 27:10445–10455
Guzowski JF et al (2006) Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc Natl Acad Sci USA 103:1077–1082
Plath N et al (2006) Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 52:437–444
Miyashita T, Kubik S, Lewandowski G, Guzowski JF (2008) A networks of neurons, networks of genes: an integrated view of memory consolidation. Neurobiol Learn Mem 89:269–284
Park S et al (2008) Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron 59:70–83
Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ (2007) The EJC factor elF4AIII modulates synaptic strength and neuronal protein expression. Cell 130:179–191
Hirokawa N, Noda Y, Tanaka Y, Niwa S (2009) Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 10:682–696
Hirokawa N (2011) From electron microscopy to molecular cell biology, molecular genetics and structural biology: intracellular transport and kinesin superfamily proteins, KIFs: genes, structure, dynamics and functions. J Electr Microsc (Tokyo) 60(Suppl 1):S63–S92
Ling SC, Fahner PS, Greenough WT, Gelfand VI (2004) Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc Natl Acad Sci USA 101:17428–17433
Dynes JL, Steward O (2007) Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J Comp Neurol 500:433–447
Nonaka S et al (1998) Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95:829–837
Jimbo T et al (2002) Identification of a link between the tumour suppressor APC and the kinesin superfamily. Nat Cell Biol 4:323–327
Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV (2003) Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426:83–87
Bananis E et al (2004) Microtubule-dependent movement of late endocytic vesicles in vitro: requirements for Dynein and Kinesin. Mol Biol Cell 15:3688–3697
Brown CL et al (2005) Kinesin-2 is a motor for late endosomes and lysosomes. Traffic 6:1114–1124
Nishimura T, Kato K, Yamaguchi T, Fukata Y, Ohno S, Kaibuchi K (2004) Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat Cell Biol 6:328–334
Shi SH, Cheng T, Jan LY, Jan YN (2004) APC and GSK-3beta are involved in mPar3 targeting to the nascent axon and establishment of neuronal polarity. Curr Biol 14:2025–2032
Haraguchi K, Hayashi T, Jimbo T, Yamamoto T, Akiyama T (2006) Role of the kinesin-2 family protein, KIF3, during mitosis. J Biol Chem 281:4094–4099
Aronov S, Aranda G, Behar L, Ginzburg I (2002) Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J Cell Sci 115:3817–3827
Kural C, Serpinskaya AS, Chou YH, Goldman RD, Gelfand VI, Selvin PR (2007) Tracking melanosomes inside a cell to study molecular motors and their interaction. Proc Natl Acad Sci USA 104:5378–5382
Watanabe TM, Higuchi H (2007) Stepwise movements in vesicle transport of HER2 by motor proteins in living cells. Biophys J 92:4109–4120
Acknowledgments
We warmly thank Carsten W. Lederer for critically proof reading the manuscript and Teresa Ciotti for her help with primary neuron cultures. D.C.C. was supported by an A.G. Leventis Foundation (France) doctoral studentship and a short-term EMBO Fellowship to visit the laboratory of C.B. for part of this work. Work in this article was included in part in the doctoral thesis of D.C.C. E.P. is supported by an FWO fellowship (aspirant). This work was funded by the University of Cyprus (N.S.), VIB and Telethon (GGP10150)HEALTH-2009-2.1.2-1 EU-FP7 ‘SynSys’ and FWO (C.B.).
Author information
Authors and Affiliations
Corresponding authors
Additional information
D. C. Charalambous, E. Pasciuto, C. Bagni, N. Santama contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2012_1108_MOESM2_ESM.eps
Specificity of the oligonucleotide pairs used for the amplification of KIF1Bβ NM015074 vs. AX039604. PCRs with the two plasmid templates (shown at the bottom) using oligonucleotides designed for NM015074 or AX039604. These oligonucleotide sets (used in panels c–g) are specific for the respective isoform in both human and mouse. Negative control was no-template PCR (neg.) (EPS 744 kb)
18_2012_1108_MOESM3_ESM.eps
Expression of KIF1B\( \alpha \) (NM008441) in mouse tissues and the NSC-34 cell line. a Schematic comparison of KIF1B\( \alpha \) and KIF1Bβ proteins, indicating regions of identity and difference in the primary structure. While the N-terminal parts of the protein are identical, more downstream sequences (starting from the point indicated by an asterisk and stretching over the sequences highlighted in gray in KIF1B1\( \alpha \)) are completely different. The oligonucleotide pair (set 3, Online Resource Table S1), hybridizing to the areas indicated on the cartoon, was used to specifically amplify KIF1B\( \alpha \). The motor (yellow) and FHA (light blue) domains are highlighted. Numbers denote amino acid (aa) positions. b Equivalent RT-PCR reactions reveal some expression of this isoform in the hippocampus and spinal motor neurons, but no expression in the mouse neuroblastoma NSC-34 line (top panels). Equivalent reactions for L19 (bottom panel). (EPS 1141 kb)
18_2012_1108_MOESM4_ESM.eps
KIF1Bβ puncta are not co-localized with markers for mitochondria, lysosomes and the Golgi but with synaptic markers. a–d. Sets of triple fluorescence images of NSC-34 cells and their overlay, showing that the punctate KIF1Bβ labeling does not co-localize with the SOD2 protein (as a marker for mitochondria) (a), or protein LAMP2 (as a marker for lysosomes) (b), or 58K Golgin (Golgi apparatus) (c), but does co-localize with a synaptic marker, post-synaptic protein PSD-95 (d). Images (from left to right): cells labeled with Hoescht (DNA), compartment-specific antibodies (as indicated), anti-KIF1Bβ, and overlay of the set of three images. Scale bars 10 μm. (EPS 3899 kb)
18_2012_1108_MOESM5_ESM.eps
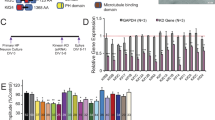
Confirmation of the effect of PMA as a stimulator of PKC. Immunoblot of cortical synaptoneurosomes that were either non-treated, or incubated with phorbol ester PMA, a stimulator of PKC, or incubated with an inhibitor of PKC. The immunoblot was probed with an antibody against the phosphorylated form of MARCKS, one of PKC substrates (top panel). GAPDH was used as internal control to demonstrate equal protein loading in the three samples (bottom panel). (EPS 1005 kb)
18_2012_1108_MOESM6_ESM.eps
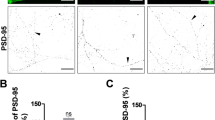
KIF1Bβ puncta are co-localized with MAP2 staining. Immunofluorescence images of cortical neurons and their overlay showing that the punctate KIF1Bβ labeling co-localizes with MAP2, a marker for dendrites. Scale bar 10 μm. A detail of the co-localization is shown at higher magnification. Scale bar 5 μm. The histogram illustrates the percentage of KIF1Bβ puncta co-localized with MAP2. (EPS 12359 kb)
18_2012_1108_MOESM7_ESM.avi
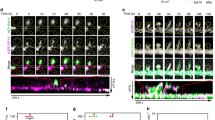
This video of a dendrite from a pEYFP-KIF1Bβ-transfected mouse cultured cortical neuron aims at providing a dynamic view to complement the still images from the live imaging shown in Fig. 8D. Fluorescent moving particles can be seen moving in the anterograde direction or in the retrograde direction or in rapid back and forth oscillating movements. Time-lapse frames were captured at a rate of one frame per 10 sec. For the analysis the time period of the first 180 sec was used. Here a movie of 100 frames, assembled with using Prism Video File Converter, is shown at 20 frames per second. (AVI 968 kb)
18_2012_1108_MOESM8_ESM.avi
Equivalent video as video 1 but derived from the pEGFP- KIF1Bβ-C1-transfected mouse cultured cortical neuron shown in Fig. 8B. Fluorescence is spread over all processes but no particle movement can be observed. (AVI 968 kb)
Rights and permissions
About this article
Cite this article
Charalambous, D.C., Pasciuto, E., Mercaldo, V. et al. KIF1Bβ transports dendritically localized mRNPs in neurons and is recruited to synapses in an activity-dependent manner. Cell. Mol. Life Sci. 70, 335–356 (2013). https://doi.org/10.1007/s00018-012-1108-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-012-1108-0