Abstract
Key message
A major QTL for SHGD was identified on chromosome 5 with stable expression across environments. The introgression this QTL can overcome the need of colchicine in DH lines development.
Abstract
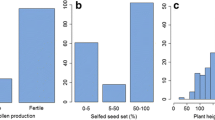
Genome doubling of haploids is one of the major constraints of large-scale doubled haploid (DH) technology. Improving spontaneous haploid genome doubling (SHGD) is an alternative to overcome this limitation. In this study, we aimed to construct a high-density linkage map based on genotyping by sequencing of single nucleotide polymorphism, to detect QTL and QTL by environment (Q by E) interactions affecting SHGD and to identify the best trait for mapping and selection of haploid male fertility (HMF). To this end, a biparental population of 220 F2:3 families was developed from a cross between A427 (high HMF) and CR1Ht (moderate HMF) to be used as donor. A high-density linkage map was constructed containing 4171 SNP markers distributed over 10 chromosomes with an average distance between adjacent markers of 0.51 cM. QTL mapping for haploid fertile anther emergence, pollen production, tassel size, and HMF, identified 27 QTL across three environments, and Q by E interactions were significant. A major QTL was identified on chromosome 5. This QTL explained over 45% of the observed variance for all traits across all environments. The introgression of this major QTL, using marker-assisted backcrossing, has great potential to overcome the need of using colchicine in DH line development.


Similar content being viewed by others
References
Beyene Y, Semagn K, Crossa J, Mugo S, Atlin GN, Tarekegne A, Meisel B, Sehabiague P, Vivek BS, Oikeh S, Alvarado G, Machida L, Olsen M, Prasanna BM, Bänziger M (2016) Improving maize grain yield under drought stress and non-stress environments in sub-Saharan Africa using marker-assisted recurrent selection. Crop Sci 56:344–353. https://doi.org/10.1007/s00122-019-03431-z
Borisy GG, Taylor EW (1967) The mechanism of action of colchicine. Binding of colchincine-3H to cellular protein. J Cell Biol 34:525–533. https://doi.org/10.1083/jcb.34.2.525
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Chaikam V, Nair SK, Martinez L, Lopez LA, Utz HF, Melchinger AE, Boddupalli PM (2018) Marker-assisted breeding of improved maternal haploid inducers in maize for the tropical/subtropical regions. Front Plant Sci 9:1527. https://doi.org/10.3389/fpls.2018.01527
Chaikam V, Gowda M, Nair SK, Melchinger AE, Boddupalli PM (2019) Genome-wide association study to identify genomic regions influencing spontaneous fertility in maize haploids. Euphytica 215:138. https://doi.org/10.1007/s10681-019-2459-5
Chalyk ST (1994) Properties of maternal haploid maize plants and potential application to maize breeding. Euphytica 79:13–18. https://doi.org/10.1007/BF00023571
Chase SS (1949) Spontaneous doubling of the chromosome complement in monoploid sporophytes of maize. Proc Iowa Acad Sci 56:15
Christensen RH (2019) Ordinal—regression models for ordinal data. Retrieved from R-project: http://www.cran.r-project.org/package=ordinal/. Accessed 26 Nov 2019
De La Fuente GN (2015) Improvements to the maize (Zea mays L.) in vivo maternal doubled haploid system. Dissertation, Iowa State University. https://doi.org/10.31274/etd-180810-4353
De la Fuente GN, Frei UK, Trampe B, Ren J, Bohn MO, Yana N et al (2020) A diallel analysis of a maize donor population response to in vivo maternal haploid induction. II. spontaneous haploid genome doubling. Crop Sci. https://doi.org/10.1002/csc2.20021
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379. https://doi.org/10.1371/journal.pone.0019379
Finkelstein Y, Aks SE, Hutson JR, Juurlink DN, Nguyen P, Dubnov-Raz G, Pollak U, Koren G, Bentur Y (2010) Colchicine poisoning: the dark side of an ancient drug. Clin Toxicol 48:407–414. https://doi.org/10.3109/15563650.2010.495348
Geiger HH, Gordillo GA (2009) Doubled haploids in hybrid maize breeding. Maydica 54:485–499
Gerdes JT, Behr CF, Coors JG, Tracy WL (1993) Compilation of North American maize breeding germplasm. Madison USA. https://doi.org/10.2135/1993.compilationofnorthamerican.frontmatter
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE 9:e90346. https://doi.org/10.1371/journal.pone.0090346
Häntzschel KR, Weber G (2010) Blockage of mitosis in maize root tips using colchicine-alternatives. Protoplasma 241:1–4. https://doi.org/10.1007/s00709-009-0103-2
Jiang L, Yang XY, Li XY, Deng PK, Jing GX, Wang XQ, Xing Z, Zhao RG (2017) A study on natural recovery of tassel fertilization and doubling method in maize haploids. Genet Mol Res 22:gmr16019173. https://doi.org/10.4238/gmr16019173
Kleiber D, Prigge V, Melchinger AE, Burkard F, San Vicente F, Palomino G, Gordillo GA (2012) Haploid fertility in temperate and tropical maize germplasm. Crop Sci 52:623. https://doi.org/10.2135/cropsci2011.07.0395
Lonnquist JH, Webel OD (1967) An evaluation of modified ear-to-row selection in a population of corn (Zea mays L.). Crop Sci 7:651–655. https://doi.org/10.2135/cropsci1967.0011183X000700060028x
Ma H, Li G, Würschum T, Zhang Y, Zheng D, Yang X, Li J, Liu W, Yan J, Chen S (2018) Genome-wide association study of haploid male fertility in maize (Zea mays L.). Front Plant Sci 9:974. https://doi.org/10.3389/fpls.2018.00974
Melchinger AE, Molenaar WS, Mirdita V, Schipprack W (2015) Colchicine alternatives for chromosome doubling in maize haploids for doubled-haploid production. Crop Sci 56:559–569. https://doi.org/10.2135/cropsci2015.06.0383
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283. https://doi.org/10.1016/j.cj.2015.01.001
Miao C, Fang J, Li D, Liang P, Zhang X, Yang J, Schnable JC, Tang H (2018) Genotype-corrector: improved genotype calls for genetic mapping in F2 and RIL populations. Sci Rep 8:10088. https://doi.org/10.1038/s41598-018-28294-0
Molenaar WS, Schipprack W, Melchinger AE (2017) Nitrous oxide-induced chromosome doubling of maize haploids. Crop Sci 58:650–659. https://doi.org/10.2135/cropsci2017.07.0412
Molenaar WS, Schipprack W, Brauner PC, Melchinger AE (2019) Haploid male fertility and spontaneous chromosome doubling evaluated in a diallel and recurrent selection experiment in maize. Theor Appl Genet 132:2273–2284. https://doi.org/10.1007/s00122-019-03353-w
Prigge V, Melchinger AE (2012) Production of haploids and doubled haploids in maize. In: Loyola-Vargas V, Ochoa-Alejo N (eds) Plant cell culture protocols. Humana Press, NJ. https://doi.org/10.1007/978-1-61779-818-4_13
R Core Team (2018) R: a language and environment for statistical computing. Vienna, Austria. http://www.R-project.org/. Accessed 16 Oct 2019
Ren J, Wu P, Tian X, Lübberstedt T, Chen S (2017) QTL mapping for haploid male fertility by a segregation distortion method and fine mapping of a key QTL qhmf4 in maize. Theor Appl Genet 130:1349–1359. https://doi.org/10.1007/s00122-017-2892-6
Ren J, Boerman N, Liu R, Wu P, Vanous K, Trampe B, Frei UK, Chen S, Lübberstedt (2019) Mapping of QTL and identification of candidate genes conferring spontaneoushaploid genome doubling in maize (Zea mays L.). Plant Sci 293:110337. https://doi.org/10.1016/j.plantsci.2019.110337
SAS Institute Inc. (2013) SAS 9.4. Cary, United States of America. https://www.sas.com/en_us/software/sas9.html. Accessed 16 Oct 2019
Schneider KL, Xie Z, Wolfgruber TK, Presting GG (2016) Inbreeding drives maize centromere evolution. PNAS USA 113:E987–E996. https://doi.org/10.1073/pnas.1522008113
Wu P, Ren J, Tian X, Lübberstedt T, Li W, Li G, Li X, Chen S (2016) New insights into the genetics of haploid male fertility in maize. Crop Sci 57:637–647. https://doi.org/10.2135/cropsci2016.01.0017
Yang J, Qu Y, Chen Q, Tang J, Lübberstedt T, Li H, Liu Z (2019) Genetic dissection of haploid male fertility in maize (Zea mays L.). Plant Breed 138:259–265. https://doi.org/10.1111/pbr.12688
Acknowledgements
This work was supported by USDA’s National Institute of Food and Agriculture (Grant Numbers: IOW04314, IOW01018, IOW05520; IOW05510; NIFA award 2018-51181-28419), the Plant Sciences Institute, Crop Bioengineering Center, R.F. Baker Center for Plant Breeding, K.J. Frey Chair in Agronomy at Iowa State University.
Author information
Authors and Affiliations
Contributions
BT, UKF, and TL design the project and performed the experiments. BT, IGS, JR, SC and TL analyzed the data. BT and IGS wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Jianbing Yan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Trampe, B., dos Santos, I.G., Frei, U.K. et al. QTL mapping of spontaneous haploid genome doubling using genotyping-by-sequencing in maize (Zea mays L.). Theor Appl Genet 133, 2131–2140 (2020). https://doi.org/10.1007/s00122-020-03585-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03585-1