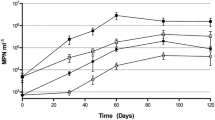
Abstract
In this report, the diversity of oil-degrading bacteria and alkB gene was surveyed in the seawater around Xiamen Island. Forty-four isolates unique in 16S rRNA sequence were obtained after enrichment with crude oil. Most of the obtained isolates exhibited growth with diesel oil and crude oil. alkB genes were positively detected in 16 isolates by degenerate polymerase chain reaction (PCR). And for the first time, alkB genes were found in bacteria of Gallaecimonas, Castellaniella, Paracoccus, and Leucobacter. Additional 29 alkB sequences were retrieved from genomic DNA of the oil-degrading communities. Phylogenetic analysis showed that the obtained alkB genes formed five groups, most of which exhibited 60–80% similarity at the amino acid level with sequences retrieved from the GenBank database. Furthermore, the abundance of alkB genes in seawater was examined by real-time PCR. The results showed that alkB genes of each group in situ ranged from about 3 × 103 to 3 × 105 copies L−1, with the homologs of Alcanivorax and Pseudomonas being the most predominant. Bacteria of Alcanivorax, Acinetobacter, and Pseudomonas are important oil degraders in this area; while those frequently reported in other area, like Oleiphilus spp., Oleispira spp., and Thalassolituus spp. were not found in our report. These results indicate that bacteria and genes involved in oil degradation are quite diverse, and may have restriction in geographic distribution in some species.



Similar content being viewed by others
References
Feng L, Wang W, Cheng J, Ren Y, Zhao G, Gao C, Tang Y, Liu X, Han W, Peng X, Liu R, Wang L (2007) Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc Natl Acad Sci USA 104:5602–5607
Golyshin PN, Martins Dos Santos VA, Kaiser O, Ferrer M, Sabirova YS, Lunsdorf H, Chernikova TN, Golyshina OV, Yakimov MM, Puhler A, Timmis KN (2003) Genome sequence completed of Alcanivorax borkumensis, a hydrocarbon-degrading bacterium that plays a global role in oil removal from marine systems. J Biotechnol 106:215–220
Head IM, Jones DM, Roling WF (2006) Marine microorganisms make a meal of oil. Nat Rev Microbiol 4:173–182
Heiss-Blanquet S, Benoit Y, Marechaux C, Monot F (2005) Assessing the role of alkane hydroxylase genotypes in environmental samples by competitive PCR. J Appl Microbiol 99:1392–1403
Islam KS, Rahman MM, Eda LEH (2009) Pollution status and sustainable management of Xiamen Bay in China: a brief review. Int J Ocean Syst Manage 1:155–168
Kuhn E, Bellicanta GS, Pellizari VH (2009) New alk genes detected in Antarctic marine sediments. Environ Microbiol 11:669–673
Kloos K, Munch JC, Schloter M (2006) A new method for the detection of alkane-monooxygenase homologous genes (alkB) in soils based on PCR-hybridization. J Microbiol Methods 66:486–496
Liu C, Shao Z (2005) Alcanivorax dieselolei sp. nov., a novel alkane-degrading bacterium isolated from sea water and deep-sea sediment. Int J Syst Evol Microbiol 55:1181–1186
Luz AP, Pellizari VH, Whyte LG, Greer CW (2004) A survey of indigenous microbial hydrocarbon degradation genes in soils from Antarctica and Brazil. Can J Microbiol 50:323–333
Maier T, Forster HH, Asperger O, Hahn U (2001) Molecular characterization of the 56-kDa CYP153 from Acinetobacter sp. EB104. Biochem Biophys Res Commun 286:652–658
Marin MM, Smits TH, van Beilen JB, Rojo F (2001) The alkane hydroxylase gene of Burkholderia cepacia RR10 is under catabolite repression control. J Bacteriol 183:4202–4209
Ou S, Zheng J, Richardson BJ, Lam PK (2004) Petroleum hydrocarbons and polycyclic aromatic hydrocarbons in the surficial sediments of Xiamen Harbour and Yuan Dan Lake, China. Chemosphere 56:107–112
Powell SM, Ferguson SH, Bowman JP, Snape I (2006) Using real-time PCR to assess changes in the hydrocarbon-degrading microbial community in Antarctic soil during bioremediation. Microb Ecol 52:523–532
Quatrini P, Scaglione G, De Pasquale C, Riela S, Puglia AM (2008) Isolation of Gram-positive n-alkane degraders from a hydrocarbon-contaminated Mediterranean shoreline. J Appl Microbiol 104:251–259
Rojo F (2009) Degradation of alkanes by bacteria. Environ Microbiol 11:2477–2490
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sei K, Sugimoto Y, Mori K, Maki H, Kohno T (2003) Monitoring of alkane-degrading bacteria in a sea-water microcosm during crude oil degradation by polymerase chain reaction based on alkane-catabolic genes. Environ Microbiol 5:517–522
Shanklin J, Whittle E, Fox BG (1994) Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochem 33:12787–12794
Siciliano SD, Fortin N, Mihoc A, Wisse G, Labelle S, Beaumier D, Ouellette D, Roy R, Whyte LG, Banks MK, Schwab P, Lee K, Greer CW (2001) Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl Environ Microbiol 67:2469–2475
Stapleton RD, Ayler GS (1998) Assessment of the microbiological potential for the natural attenuation of petroleum hydrocarbons in a shallow aquifer system. Microbial Ecol: 36:49–361
Smits TH, Röthlisberger M, Witholt B, van Beilen JB (1999) Molecular screening for alkane hydroxylase genes in gram-negative and gram-positive strains. Environ Microbiol 1:307–317
Tani A, Ishige T, Sakai Y, Kato N (2001) Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1. J Bacteriol 183:1819–1823
Throne-Holst M, Wentzel A, Ellingsen TE, Kotlar HK, Zotchev SB (2007) Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl Environ Microbiol 73:3327–3332
van Beilen JB, Wubbolts MG, Witholt B (1994) Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5:161–174
van Beilen JB, Smits THM, Whyte LG, Schorcht S, Röthlisberger M, Plaggemeier T, Engesser KH, Witholt B (2002) Alkane hydroxylases in gram-positive strains. Environ Microbiol 4:676–682
van Beilen JB, Duetz WA, Schmid A, Witholt B (2003) Practical issues in the application of oxygenases. Trends Biotechnol 21:170–177
van Beilen JB, Li Z, Duetz WA, Smits THM, Witholt B (2003) Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci Technol 58:427–440
van Beilen JB, Funhoff EG (2005) Expanding the alkane oxygenase toolbox: new enzymes and applications. Curr Opin Biotechnol 16:308–314
van Beilen JB, Funhoff EG (2007) Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol 74:13–21
Vomberg A, Klinner U (2000) Distribution of alkB genes within n alkane-degrading bacteria. J Appl Microbiol 89:339–348
Wang L, Wang W, Lai Q, Shao Z (2010) Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean. Environ Microbiol 12(5):1230–1242
Wasmund K, Burns KA, Kurtboke DI, Bourne DG (2009) Novel alkane hydroxylase gene (alkB) diversity in sediments associated with hydrocarbon seeps in the Timor Sea, Australia. Appl Environ Microbiol 75:7391–7398
WEF AA (1998) Standard Methods for the Examination of Water and Wastewater (20th ed.), APHA-AWWA-WEF, Washington, DC
Wentzel A, Ellingsen TE, Kotlar HK, Zotchev SB, Throne-Holst M (2007) Bacterial metabolism of long-chain n-alkanes. Appl Microbiol Biotechnol 76:1209–1221
Whyte LG, Schultz A, Van Beilen JB, Luz AP, Pellizari D, Labbé D, Greer CW (2002) Prevalence of alkane monooxygenase genes in arctic and antarctic hydrocarbon-contaminated and pristine soils. FEMS Microbiol Ecol 41:141–150
Whyte LG, Smits TH, Labbe D, Witholt B, Greer CW, van Beilen JB (2002) Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl Environ Microbiol 68:5933–5942
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (30670051), the Science and Technology Program of Fujian Province of China (2009H0029), and the China Scholarship Council Program (2008631036).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Figure S1
Multiple alignment of each alkB group to show the conserved regions used for Q-PCR primer design. Q-PCR primers of each group were designed based on the two conserved regions labeled in grey pane. a. Group I-like alkB gene; b. Group II-like alkB gene; c. Group III-like alkB gene; d. Group IV-like alkB gene; e. Group V-like alkB gene. (GIF 750 kb)
Rights and permissions
About this article
Cite this article
Wang, W., Wang, L. & Shao, Z. Diversity and Abundance of Oil-Degrading Bacteria and Alkane Hydroxylase (alkB) Genes in the Subtropical Seawater of Xiamen Island. Microb Ecol 60, 429–439 (2010). https://doi.org/10.1007/s00248-010-9724-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-010-9724-4