Abstract
Bats are important zoonotic reservoirs for many pathogens worldwide. Although their highly specialized ectoparasites, bat flies (Diptera: Hippoboscoidea), can transmit Bartonella bacteria including human pathogens, their eco-epidemiology is unexplored. Here, we analyzed the prevalence and diversity of Bartonella strains sampled from 10 bat fly species from 14 European bat species. We found high prevalence of Bartonella spp. in most bat fly species with wide geographical distribution. Bat species explained most of the variance in Bartonella distribution with the highest prevalence of infected flies recorded in species living in dense groups exclusively in caves. Bat gender but not bat fly gender was also an important factor with the more mobile male bats giving more opportunity for the ectoparasites to access several host individuals. We detected high diversity of Bartonella strains (18 sequences, 7 genotypes, in 9 bat fly species) comparable with tropical assemblages of bat-bat fly association. Most genotypes are novel (15 out of 18 recorded strains have a similarity of 92–99%, with three sequences having 100% similarity to Bartonella spp. sequences deposited in GenBank) with currently unknown pathogenicity; however, 4 of these sequences are similar (up to 92% sequence similarity) to Bartonella spp. with known zoonotic potential. The high prevalence and diversity of Bartonella spp. suggests a long shared evolution of these bacteria with bat flies and bats providing excellent study targets for the eco-epidemiology of host-vector-pathogen cycles.








Similar content being viewed by others
References
Tsang SM, Cirranello AL, Bates PJ, Simmons NB (2015) The roles of taxonomy and systematics in bat conservation. In: Voigt CC, Kingston T (eds) Bats in the Anthropocene: conservation of bats in a changing world. Springer International Publishing, pp 503–538
Teeling EC (2009) A molecular phylogeny for bats illuminates biogeography and the fossil record a molecular phylogeny for bats illuminates biogeography and the fossil record. Notes 307:580–584. https://doi.org/10.1126/science.1105113
Haelewaters D, Pfliegler WP, Szentiványi T, Földvári M, Sándor AD, Barti L, Camacho JJ, Gort G, Estók P, Hiller T, Dick CW, Pfister DH (2017) Parasites of parasites of bats: Laboulbeniales (Fungi: Ascomycota) on bat flies (Diptera: Nycteribiidae) in Central Europe. Parasit. Vectors 10:96. https://doi.org/10.1186/s13071-017-2022-y
Dick CW, Patterson BD (2006) Bat flies-obligate ectoparasites of bats. In: Morand S, Krasnov BR, RP (eds) Micromammals and macroparasites: from evolutionary ecology to management. Springer-Verlag, Tokyo Berlin Heidelberg New York, pp 179–194
Dick CW, Dittmar K (2014) Parasitic bat flies (Diptera: Streblidae and Nycteribiidae): host specificity and potential as vectors. In: Klimpel SMH (ed) Bats (Chiroptera) as vectors of diseases and parasites. Springer-Verlag, Berlin Heidelberg, pp 131–151
Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, Bryden WL, Middleton D, Reid PA, McFarlane RA, Martin G, Tabor GM, Skerratt LF, Anderson DL, Crameri G, Quammen D, Jordan D, Freeman P, Wang LF, Epstein JH, Marsh GA, Kung NY, McCallum H (2015) Ecological dynamics of emerging bat virus spillover. Proc. R. Soc. B Biol. Sci. 282:20142124. https://doi.org/10.1098/rspb.2014.2124
Mühldorfer K (2013) Bats and bacterial pathogens: a review. Zoonoses Public Health 60:93–103. https://doi.org/10.1111/j.1863-2378.2012.01536.x
Hornok S, Szőke K, Kováts D, Estók P, Görföl T, Boldogh SA, Takács N, Kontschán J, Földvári G, Barti L, Corduneanu A, Sándor AD (2016) DNA of piroplasms of ruminants and dogs in ixodid bat ticks. PLoS One 11:e0167735. https://doi.org/10.1371/journal.pone.0167735
Corduneanu A, Hrazdilová K, Sándor AD, et al (2017) Babesia vesperuginis, a neglected piroplasmid: new host and geographical records, and phylogenetic relations. Parasit. Vectors 10:598. https://doi.org/10.1186/s13071-017-2424-x
Hornok S, Szőke K, Tu VT, Kontschán J, Takács N, Sándor AD, Halajian A, Földvári G, Estók P, Plantard O, Epis S, Görföl T (2017) Mitochondrial gene heterogeneity of the bat soft tick Argas vespertilionis (Ixodida: Argasidae) in the Palaearctic. Parasit. Vectors 10:109. https://doi.org/10.1186/s13071-017-2037-4
Gibson KE, Rikihisa Y, Zhang C, Martin C (2005) Neorickettsia risticii is vertically transmitted in the trematode Acanthatrium oregonense and horizontally transmitted to bats. Environ. Microbiol. 7:203–212. https://doi.org/10.1111/j.1462-2920.2004.00683.x
Evans NJ, Bown K, Timofte D, Simpson VR, Birtles RJ (2009) Fatal borreliosis in bat caused by relapsing fever spirochete, United Kingdom. Emerg. Infect. Dis. 15:1331–1333
Hosokawa T, Nikoh N, Koga R, Satô M, Tanahashi M, Meng XY, Fukatsu T (2012) Reductive genome evolution, host–symbiont co-speciation and uterine transmission of endosymbiotic bacteria in bat flies. ISME J 6:577–587. https://doi.org/10.1038/ismej.2011.125
Lei BR, Olival KJ (2014) Contrasting patterns in mammal-bacteria coevolution: Bartonella and Leptospira in bats and rodents. PLoS Negl. Trop. Dis. 8:e2738. https://doi.org/10.1371/journal.pntd.0002738
Hanson A (1970) Isolation of spirochaetes from primates and other mammalian species. Br J Vener Dis 46:303–306. https://doi.org/10.1136/sti.46.4.303
Concannon R, Wynn-Owen K, Simpson VR, Birtles RJ (2005) Molecular characterization of haemoparasites infecting bats (Microchiroptera) in Cornwall, UK. Parasitology 131:489–496. https://doi.org/10.1017/S0031182005008097
Segers FH, Kešnerová L, Kosoy M, Engel P (2017) Genomic changes associated with the evolutionary transition of an insect gut symbiont into a blood-borne pathogen. ISME J 11:1232–1244. https://doi.org/10.1038/ismej.2016.201
Boulouis HJ, Chang CC, Henn JB et al (2005) Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet. Res. 36:383–410. https://doi.org/10.1051/vetres:2005009
Chomel BB, Boulouis HJ, Breitschwerdt EB, Kasten RW, Vayssier-Taussat M, Birtles RJ, Koehler JE, Dehio C (2009) Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet. Res. 40:29. https://doi.org/10.1051/vetres/2009011
Breitschwerdt EB (2014) Bartonellosis: one health perspectives for an emerging infectious disease. ILAR J. 55:46–58. https://doi.org/10.1093/ilar/ilu015
Kosoy M, Bai Y, Lynch T, Kuzmin IV, Niezgoda M, Franka R, Agwanda B, Breiman RF, Rupprecht CE (2010) Bartonella spp. in bats, Kenya. Emerg. Infect. Dis. 16:1875–1881. https://doi.org/10.3201/eid1612.100601
Bai Y, Hayman DTS, McKee CD, Kosoy MY (2015) Classification of Bartonella strains associated with straw-colored fruit bats (Eidolon helvum) across Africa using a multi-locus sequence typing platform. PLoS Negl. Trop. Dis. 9:e0003478. https://doi.org/10.1371/journal.pntd.0003478
Wilkinson DA, Duron O, Cordonin C, Gomard Y, Ramasindrazana B, Mavingui P, Goodman SM, Tortosa P (2016) The bacteriome of bat flies (Nycteribiidae) from the Malagasy region: a community shaped by host ecology, bacterial transmission mode, and host-vector specificity. Appl. Environ. Microbiol. 82:1778–1788. https://doi.org/10.1128/AEM.03505-15
Anh PH, Van Cuong N, Son NT et al (2015) Diversity of bartonella spp. in bats, southern Vietnam. Emerg. Infect. Dis. 21:1266–1267. https://doi.org/10.3201/eid2107.141760
Han HJ, Wen HL, Zhao L, Liu JW, Luo LM, Zhou CM, Qin XR, Zhu YL, Zheng XX, Yu XJ (2017) Novel Bartonella species in insectivorous bats, Northern China. PLoS One 12:e0167915. https://doi.org/10.1371/journal.pone.0167915
Olival KJ, Dittmar K, Bai Y, Rostal MK, Lei BR, Daszak P, Kosoy M (2015) Bartonella spp. in a Puerto Rican bat community. J. Wildl. Dis. 51:274–278. https://doi.org/10.7589/2014-04-113
McKee CD, Hayman DTS, Kosoy MY, Webb CT (2016) Phylogenetic and geographic patterns of bartonella host shifts among bat species. Infect Genet Evol 44:382–394. https://doi.org/10.1016/j.meegid.2016.07.033
Hornok S, Kovács R, Meli ML, Gönczi E, Hofmann-Lehmann R, Kontschán J, Gyuranecz M, Dán Á, Molnár V (2012) First detection of bartonellae in a broad range of bat ectoparasites. Vet. Microbiol. 159:541–543. https://doi.org/10.1016/j.vetmic.2012.04.003
Veikkolainen V, Vesterinen EJ, Lilley TM, Pulliainen AT (2014) Bats as reservoir hosts of human bacterial pathogen, Bartonella mayotimonensis. Emerg. Infect. Dis. 20:960–967. https://doi.org/10.3201/eid2006.130956
Haysom K, Dekker J, Russ J et al (2013) European bat population trends. A prototype biodiversity indicator, Technical. European Environment Agency, Bruxelles
Dietz C, von Helversen O, Nill D (2009) Bats of Britain, Europe and Northwest Africa. A & C Black, London
Theodor O (1967) An illustrated catalogue of the Rothschild collection of Nycteribiidae in the British Museum (Natural History), with keys and short descriptions for the identification of subfamilies, genera, species and subspecies. British Museum (Natural History) Publication 655, London
Theodor O, Moscona A (1954) On bat parasites in Palestine. I. Nycteribiidae, Streblidae, Hemiptera, Siphonaptera. Parasitology 44:157–245. https://doi.org/10.1017/S0031182000018862
Wielinga PR, Gaasenbeek C, Fonville M, de Boer A, de Vries A, Dimmers W, Akkerhuis op Jagers G, Schouls LM, Borgsteede F, van der Giessen JWB (2006) Longitudinal analysis of tick densities and Borrelia, Anaplasma, and Ehrlichia infections of Ixodes ricinus ticks in different habitat areas in the Netherlands. Appl. Environ. Microbiol. 72:7594–7601. https://doi.org/10.1128/AEM.01851-06
Norman a F, Regnery R, Jameson P et al (1995) Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 33:1797–1803
Dormann CF, Frund J, Bluthgen N, Gruber B (2009) Indices, graphs and null models: analyzing bipartite ecological networks. Open Ecol J 2:7–24. https://doi.org/10.2174/1874213000902010007
Core Team R (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Viena
Bates D, Maechler M, Bolker B, Walker S (2015) lm4: Linear mixded-effects models using Eigen and S4
Morse SF, Dick CW, Patterson BD, Dittmard K (2012) Some like it hot: evolution and ecology of novel endosymbionts in bat flies of cave-roosting bats (Hippoboscoidea, Nycterophiliinae). Appl. Environ. Microbiol. 78:8639–8649. https://doi.org/10.1128/AEM.02455-12
Morse SF, Olival KJ, Kosoy M et al (2012) Global distribution and genetic diversity of Bartonella in bat flies (Hippoboscoidea, Streblidae, Nycteribiidae). Infect Genet Evol 12:1717–1723. https://doi.org/10.1016/j.meegid.2012.06.009
Brook CE, Bai Y, Dobson AP, Osikowicz LM, Ranaivoson HC, Zhu Q, Kosoy MY, Dittmar K (2015) Bartonella spp. in fruit bats and blood-feeding ectoparasites in Madagascar. PLoS Negl. Trop. Dis. 10:e0003532. https://doi.org/10.1371/journal.pntd.0003532
Billeter S a, Hayman DTS, Peel aJ et al (2012) Bartonella species in bat flies (Diptera: Nycteribiidae) from western Africa. Parasitology 139:324–329. https://doi.org/10.1017/S0031182011002113
Judson SD, Frank HK, Hadly EA (2015) Bartonellae are prevalent and diverse in Costa Rican bats and bat flies. Zoonoses Public Health 62:609–617. https://doi.org/10.1111/zph.12188
Dietrich M, Tjale MA, Weyer J, Kearney T, Seamark ECJ, Nel LH, Monadjem A, Markotter W (2016) Diversity of bartonella and rickettsia spp. in bats and their blood-feeding Ectoparasites from South Africa and Swaziland. PLoS One 11:e0152077. https://doi.org/10.1371/journal.pone.0152077
Stuckey MJ, Boulouis HJ, Cliquet F, Picard-Meyer E, Servat A, Aréchiga-Ceballos N, Echevarría JE, Chomel BB (2017) Potentially zoonotic bartonella in bats from France and Spain. Emerg. Infect. Dis. 23:539–541. https://doi.org/10.3201/eid2303.160934
Szentiványi T, Estók P, Földvári M (2016) Checklist of host associations of European bat flies (Diptera: Nycteribiidae, Streblidae). Zootaxa 4205:101–126. https://doi.org/10.11646/zootaxa.4205.2.1
Gutiérrez R, Morick D, Cohen C, Hawlena H, Harrus S (2014) The effect of ecological and temporal factors on the composition of Bartonella infection in rodents and their fleas. ISME J 8:1598–1608. https://doi.org/10.1038/ismej.2014.22
Marshall AG (1982) Ecology of insects ectoparasitic on bats. In: Kunz TE (ed) Ecology of bats. Springer, US, Philadelphia, pp 369–401
Petit E, Mayer F (1999) Male dispersal in the noctule bat (Nyctalus noctula): where are the limits? Proc. Biol. Sci. 266:1717–1722. https://doi.org/10.1098/rspb.1999.0837
Encarnacao J a, Kierdorf U, Wolters V (2006) Seasonal variation in nocturnal activity of male Daubenton ’ s bats , Myotis daubentonii ( Chiroptera : Vespertilionidae). Folia Zool. 55:237–246
Moussy C, Hosken DJ, Mathews F, Smith GC, Aegerter JN, Bearhop S (2013) Migration and dispersal patterns of bats and their influence on genetic structure. Mamm Rev 43:183–195. https://doi.org/10.1111/j.1365-2907.2012.00218.x
Ramos Pereira MJ, Salgueiro P, Rodrigues L, Coelho MM, Palmeirim JM (2009) Population structure of a cave-dwelling bat, Miniopterus schreibersii: does it reflect history and social organization? J Hered 100:533–544. https://doi.org/10.1093/jhered/esp032
Altringham JD, Senior P, Ruckstuhl KE, Neuhaus P (2005) Social systems and ecology of bats. In: Ruckstuhl KE, Neuhaus P (eds) Sexual segregation in vertebrates. Cambridge University Press, Cambridge, pp 280–302
Rodrigues L, Palmeirim JM (2008) Migratory behaviour of the Schreiber’s bat: when, where and why do cave bats migrate in a Mediterranean region? J. Zool. 274:116–125. https://doi.org/10.1111/j.1469-7998.2007.00361.x
Gutiérrez R, Vayssier-Taussat M, Buffet J-P, Harrus S (2017) Guidelines for the isolation, molecular detection, and characterization of Bartonella species. Vector-Borne Zoonotic Dis 17:42–50. https://doi.org/10.1089/vbz.2016.1956
Kosoy M, Morway C, Sheff KW, Bai Y, Colborn J, Chalcraft L, Dowell SF, Peruski LF, Maloney SA, Baggett H, Sutthirattana S, Sidhirat A, Maruyama S, Kabeya H, Chomel BB, Kasten R, Popov V, Robinson J, Kruglov A, Petersen LR (2008) Bartonella tamiae sp. nov., a newly recognized pathogen isolated from three human patients from Thailand. J. Clin. Microbiol. 46:772–775. https://doi.org/10.1128/JCM.02120-07
Acknowledgements
Permission for bat capture was provided by the National Inspectorate for Environment, Nature and Water (Hungary), and the Underground Heritage Commission (Romania). Bat banding license numbers are 59/2003 (PE), 305/2015 (ADS), and TMF-493/3/2005 (TG). No live bat was harmed for this study. The authors thank C. Jére, I. Csősz, D. Bălășoiu, A. Telea, and A. Ionică for their contribution in the bat fly collection in Romania. We would like to express our thanks to Cristian Domșa for the help provided in the creation of the maps in Figs. 1 and 6. The survey was organized in the framework of the EurNegVec COST Action TD1303. This research was supported from the grant PN-II-RU-TE-2014-4-1389, the grant “In the light of evolution: theories and solutions” (GINOP-2.3.2-15-2016-00057), and the János Bolyai Research Scholarship of the Hungarian Academy of Science (to ADS and GF).
Author information
Authors and Affiliations
Contributions
ADS initiated the study, did part of the sample collection, and wrote the manuscript. LB, AC, SH, TG, PE, ZL, and DK contributed important samples to the study. MF identified all bat flies. AK, SSz, and HS performed the molecular and phylogenetic analyses. GF organized part of the sample collection and contributed to the study design and manuscript preparation. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Figure S1
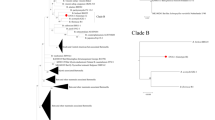
Neighbor-joining phylogenetic tree based on the multiple alignment of the gltaA gene, including Bartonella sequences obtained in this study. Clusters were assigned based on visual inspection of the tree. (PDF 65 kb)
Supplementary Figure S2
Neighbor-joining phylogenetic tree based on the pairwise alignment of an approx. 360 bp long fragment of the gltaA gene, including representative Bartonella sequences obtained in this study (highlighted by green fonts and 5 character codes) and representative sequences of other Bartonella spp. retrieved from GenBank. Sequences in blue represent Bartonella spp. related to bats or bat flies, while sequences in black denote Bartonella spp. from other host species, Bartonella spp. with known pathogenicity are highlighted in bold. (PDF 31 kb)
Table S1
(XLSX 11 kb)
Table S2
(XLSX 35 kb)
Table S3
(XLSX 10 kb)
Table S4
(DOCX 15 kb)
Table S5
(XLSX 10 kb)
Rights and permissions
About this article
Cite this article
Sándor, A.D., Földvári, M., Krawczyk, A.I. et al. Eco-epidemiology of Novel Bartonella Genotypes from Parasitic Flies of Insectivorous Bats. Microb Ecol 76, 1076–1088 (2018). https://doi.org/10.1007/s00248-018-1195-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-018-1195-z