Abstract
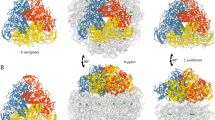
Ribose-5-phosphate isomerase (Rpi) catalyzes the conversion of d-ribose 5-phosphate (R5P) to d-ribulose 5-phosphate, which is an important step in the non-oxidative pathway of the pentose phosphate pathway and the Calvin cycle of photosynthesis. Recently, Rpis have been used to produce valuable rare sugars for industrial purposes. Of the Rpis, d-ribose-5-phosphate isomerase B from Clostridium thermocellum (CtRpi) has the fastest reactions kinetics. While Thermotoga maritime Rpi (TmRpi) has the same substrate specificity as CtRpi, the overall activity of CtRpi is approximately 200-fold higher than that of TmRpi. To understand the structural basis of these kinetic differences, we determined the crystal structures, at 2.1-Å resolution or higher, of CtRpi alone and bound to its substrates, R5P, d-ribose, and d-allose. Structural comparisons of CtRpi and TmRpi showed overall conservation of their structures with two notable differences. First, the volume of the CtRpi substrate binding pocket (SBP) was 20% less than that of the TmRpi SBP. Second, the residues next to the sugar-ring opening catalytic residue (His98) were different. We switched the key residues, involved in SBP shaping or catalysis, between CtRpi and TmRpi by site-directed mutagenesis, and studied the enzyme kinetics of the mutants. We found that tight interactions between the two monomers, narrow SBP width, and the residues near the catalytic residue are all critical for the fast enzyme kinetics of CtRpi.





Similar content being viewed by others
References
Abagyan R, Totrov M, Kuznetsov D (1994) ICM—a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comput Chem 15:488–506
Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132
Graille M, Meyer P, Leulliot N, Sorel I, Janin J, Van Tilbeurgh H, Quevillon-Cheruel S (2005) Crystal structure of the S. cerevisiae D-ribose 5-phosphate isomerase: comparison with the archaeal and bacterial enzymes. Biochimie 87:763–769
Holmes MA, Buckner FS, Van Voorhis WC, Verlinde CL, Mehlin C, Boni E, DeTitta G, Luft J, Lauricella A, Anderson L, Kalyuzhniy O, Zucker F, Schoenfeld LW, Earnest TN, Hol WG, Merritt EA (2006) Structure of ribose 5-phosphate isomerase from Plasmodium falciparum. Acta Crystallogr F Struct Biol Cryst Commun 62:427–431
Hossain MA, Wakabayashi H, Goda F, Kobayashi S, Maeba T, Maeta H (2000) Effect of the immunosuppressants FK506 and D-allose on allogenic orthotopic liver transplantation in rats. Transplant Proc 32:2021–2023
Hossain MA, Izuishi K, Tokuda M, Izumori K, Maeta H (2004) D-Allose has a strong suppressive effect against ischemia/reperfusion injury: a comparative study with allopurinol and superoxide dismutase. J Hepatobiliary Pancreat Surg 11:181–189
Ishikawa K, Matsui I, Payan F, Cambillau C, Ishida H, Kawarabayasi Y, Kikuchi H, Roussel A (2002) A hyperthermostable D-ribose 5-phosphate isomerase from Pyrococcus horikoshii characterization and three-dimensional structure. Structure 10:877–886
Kleywegt GJ (2007) Crystallographic refinement of ligand complexes. Acta Crystallogr D Biol Crystallogr 63:94–100
Levin GV (2002) Tagatose, the new GRAS sweetener and health product. J Med Food 5:23–36
Levin GV, Zehner LR, Saunders JP, Beadle JR (1995) Sugar substitutes: their energy values, bulk characteristics, and potential health benefits. Am J Clin Nutr 62:1161S–1168S
Livesey G, Brown JC (1996) D-tagatose is a bulk sweetener with zero energy determined in rats. J Nutr 126:1601–1609
Mathews BW (1968) Solvent content of protein crystals. J Mol Biol 32:491–497
Matsuo T, Suzuki H, Hashiguchi M, Izumori K (2002) D-psicose is a rare sugar that provides no energy to growing rats. J Nutr Sci Vitaminol (Tokyo) 48:77–80
Muniruzzaman S, Pan YT, Zeng Y, Atkins B, Izumori K, Elbein AD (1996) Inhibition of glycoprotein processing by L-fructose and L-xylulose. Glycobiology 6:795–803
Murata A, Sekiya K, Watanabe Y, Yamaguchi F, Hatano N, Izumori K, Tokuda M (2003) A novel inhibitory effect of D-allose on production of reactive oxygen species from neutrophils. J Biosci Bioeng 96:89–91
Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255
Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 277:307–326
Poulsen TS, Chang YY, Hove-Jensen B (1999) D-Allose catabolism of Escherichia coli: involvement of alsI and regulation of als regulon expression by allose and ribose. J Bacteriol 181:7126–7130
Rangarajan ES, Sivaraman J, Matte A, Cygler M (2002) Crystal structure of D-ribose 5-phosphate isomerase (RpiA) from Escherichia coli. Proteins 48:737–740
Roos AK, Andersson CE, Bergfors T, Jacobsson M, Karlen A, Unge T, Jones TA, Mowbray SL (2004) Mycobacterium tuberculosis ribose-5-phosphate isomerase has a known fold, but a novel active site. J Mol Biol 335:799–809
Roos AK, Mariano S, Kowalinski E, Salmon L, Mowbray SL (2008) D-ribose 5-phosphate isomerase B from Escherichia coli is also a functional D-allose 6-phosphate isomerase, while the Mycobacterium tuberculosis enzyme is not. J Mol Biol 382:667–679
Strange RW, Antonyuk SV, Ellis MJ, Bessho Y, Kuramitsu S, Yokoyama S, Hasnain SS (2009) The structure of an archaeal ribose 5-phosphate isomerase from Methanocaldococcus jannaschii (MJ1603). Acta Crystallogr F Struct Biol Cryst Commun 65:1214–1217
Xu Q, Schwarzenbacher R, McMullan D, von Delft F, Brinen LS, Canaves JM, Dai X, Deacon AM, Elsliger MA, Eshagi S, Floyd R, Godzik A, Grittini C, Grzechnik SK, Jaroszewski L, Karlak C, Klock HE, Koesema E, Kovarik JS, Kreusch A, Kuhn P, Lesley SA, Levin I, McPhillips TM, Miller MD, Morse A, Moy K, Ouyang J, Page R, Quijano K, Robb A, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, Vincent J, Wang X, West B, Wolf G, Hodgson KO, Wooley J, Wilson IA (2004) Crystal structure of a ribose-5-phosphate isomerase RpiB (TM1080) from Thermotoga maritima at 1.90 Å resolution. Proteins 56:171–175
Yeom SJ, Kim BN, Park CS, Oh DK (2010) Substrate specificity of ribose-5-phosphate isomerases from Clostridium difficile and Thermotoga maritima. Biotechnol Lett 32:829–835
Yoon RY, Yeom SJ, Kim HJ, Oh DK (2009) Novel substrates of a ribose-5-phosphate isomerase from Clostridium thermocellum. J Biotechnol 139:26–32
Zhang RG, Andersson CE, Savchenko A, Skarina T, Evdokimova E, Beasley S, Arrowsmith CH, Edwards AM, Joachimiak A, Mowbray SL (2003a) Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle. Structure 11:31–42
Zhang RG, Andersson CE, Skarina T, Evdokimova E, Edwards AM, Joachimiak A, Savchenko A, Mowbray SL (2003b) The 2.2 Å resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction. J Mol Biol 332:1083–1094
Acknowledgments
We are grateful to Dr. Y. G. Kim and Dr. K. J. Kim for their assistance at the beamline 4A and 6C of Pohang Light Source (PLS) in South Korea, and to staff members at the beamline 17A of the photon factory (KEK), Japan. This work was supported by a WCU (World Class University, R33-2008-000-1071) program through the Korea Science and Engineering Foundation funded by the Ministry of Education, Science and Technology, South Korea; by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0011666); and by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST, No. 2010-0027545).
Accession numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 3HE8 (Apo-CtRpi), 3HEE (CtRpi-R5P), 3PH3 (CtRpi-RBO), and 3PH4 (CtRpi-ALO).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Junho Jung and Jin-Kwang Kim equally contributed to this paper.
Rights and permissions
About this article
Cite this article
Jung, J., Kim, JK., Yeom, SJ. et al. Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics. Appl Microbiol Biotechnol 90, 517–527 (2011). https://doi.org/10.1007/s00253-011-3095-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3095-8