Abstract
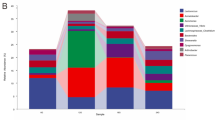
Coilia nasus is influenced by various external pressures during spawning migration and these anadromous transitions can lead to specific gut microbiome characteristics that affecting the host biological process. Therefore, the purpose of this study was to determine the variations of components and functions in the gut prokaryotic microbiome during spawning migration as well as the key factors that triggered the changes. The gut microbiome in C. nasus was mainly consisted of Proteobacteria, Bacteroidetes, Firmicutes, Deinococcus-Thermus and Fusobacteria via 16S rRNA Gene Amplicon Sequencing. The relative abundance of Acinetobacter and Clostridium increased, while Corynebacterium, Actinomyces, Bacillus, Klebsiella and Ochrobactrum decreased after entering freshwater, indicated the preference of C. nasus gut microbial members transferred from seawater to freshwater. Additionally, the proportion of Firmicutes significantly decreased and then increased, as well as the arise of some soil bacteria in gut, corresponding to the phenomenon that C. nasus are fasting during the upstream process and refeeding after entering the spawning grounds. The function prediction of gut microbiome was also consistent with the above results. The present study generally demonstrated the gut microbiome dynamics and the significant correlation between the gut microbiome and salinity and feeding behavior in the spawning migration of C. nasus.





Similar content being viewed by others
References
Wang M, Xu P, Zhu Z (2020) Regulation of signal transduction in Coilia nasus during migration. Genomics 112(1):55–64
Zhu G, Wang L, Tang W et al (2014) De novo transcriptomes of olfactory epithelium reveal the genes and pathways for spawning migration in Japanese grenadier anchovy (Coilia nasus). PLoS ONE. https://doi.org/10.1371/journal.pone.0103832
Feng XT, Yang XJ, Ruan JJ et al (2019) Molecular cloning and characteristics of DnaJa1and DnaJb1 in Coilia nasus: possible function involved in oogenesis during spawning migration. BMC Dev Biol. https://doi.org/10.1186/s12861-019-0187-7
Song R, Li WX, Wu SG et al (2014) Population genetic structure of the acanthocephalan acanthosentis cheni in anadromous, freshwater, and landlocked stocks of its fish host, Coilia nasus. J Parasitol 100(2):193–197
Ma FJ, Yang YP, Jiang M et al (2019) Digestive enzyme activity of the Japanese grenadier anchovy Coilia nasus during spawning migration: influence of the migration distance and the water temperature. J Fish Biol 95(5):1311–1319
Jiang T, Yang J, Liu H et al (2012) Life history of Coilia nasus from the yellow sea inferred from otolith Sr: Ca ratios. Environ Biol Fish 95(4):503–508
Xu JB, Deng PP, Shi YH et al (2016) Effect of salinity on the survival and growth of the larvae and juveniles of japanese grenadier anchovy Coilia nasus. N Am J Aouacult 78(1):1–7
Nie ZJ, Xu GC, Du FK et al (2015) PCR-DGGE fingerprinting and diversity analysis of the predominant bacterial community in Coilia nasus. Acta Hydrobiol Sin 39(5):1019–1026
Dimitroglou A, Merrifield DL, Carnevali O et al (2011) Microbial manipulations to improve fish health and production—a Mediterranean perspective. Fish Shellfish Immun 30(1):1–16
Li XM, Yu YH, Feng WS et al (2012) Host species as a strong determinant of the intestinal microbiota of fish larvae. J Microbiol 50:29–37
Li XM, Yan QY, Ringø E et al (2016) The influence of weight and gender on intestinal bacterial community of wild largemouth bronze gudgeon (Coreius guichenoti, 1874). BMC Microbiol. https://doi.org/10.1186/s12866-016-0809-1
Stephens WZ, Burns AR, Stagaman K et al (2016) The composition of the zebrafish intestinal microbial community varies across development. ISME J 10:644–654
Bolnick DI, Snowberg LK, Caporaso JG et al (2014) Major histocompatibility complex class IIb polymorphism influences gut microbiota composition and diversity. Mol Ecol 23(19):4831–4845
Martin SL, Philip M, Melanie D et al (2016) The biogeography of the Atlantic salmon (Salmo salar) gut microbiome. ISME J 10(5):1280–1284
Hovda MB, Fontanillas R, McGurk C et al (2012) Seasonal variations in the intestinal microbiota of farmed Atlantic salmon (Salmo salar L.). Aquac Res 43:154–159
Vasemägi A, Visse M, Kisand V (2017) Effect of environmental factors and an emerging parasitic disease on gut microbiome of wild salmonid fish. Msphere. https://doi.org/10.1128/mSphere.00418-17
Ringø E, Zhou Z, Vecino JLG et al (2016) Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquac Nutr 22:219–282
Dehler CE, Secombes CJ, Martin SAM (2017) Seawater transfer alters the intestinal microbiota profiles of Atlantic salmon (Salmo salar L.). Sci Rep. https://doi.org/10.1038/s41598-017-13249-8
Knut R, Angell IL, Pope PB et al (2018) Stable core gut microbiota across the freshwater-to-saltwater transition for farmed Atlantic Salmon. Appl Environ Microb. https://doi.org/10.1128/AEM.01974-17
Lozupone CA, Stombaugh J, Gonzalez A et al (2013) Meta-analyses of studies of the human microbiota. Genome Res 23(10):1704–1714
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Desantis TZ, Hugenholtz P, Larsen N et al (2006) Greengenes, a Chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb 72(7):5069–5072
Lozupone CA, Hamady M, Kelley ST et al (2007) Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microb 73(5):1576–1585
Ramette A (2007) Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62(2):142–160
Mcardle BH, Anderson MJ (2001) Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82(1):290–297
Warton DI, Wright ST, Wang Y (2012) Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol Evol 3(1):89–101
Zaura E, Keijser BJ, Huse SM et al (2009) Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol. https://doi.org/10.1186/1471-2180-9-259
White JR, Nagarajan N, Pop M (2009) Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1000352
Langille MG, Zaneveld J, Caporaso JG et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31(9):814–821
Parks DH, Tyson GW, Hugenholtz P et al (2014) STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30(21):3123–3124
Logares R, Bråte J, Bertilsson S et al (2009) Infrequent marine–freshwater transitions in the microbial world. Trends Microbiol. https://doi.org/10.1016/j.tim.2009.05.010
Li WX, Rui S, Wu SG et al (2011) Seasonal occurrence of helminths in the anadromous fish Coilia nasus (Engraulidae): parasite indicators of fish migratory movements. J Parasitol 97(2):192–196
Guan W, Chen H (2014) Energy dynamics in anadromous C. Ectens during spawning and overwintering in Yangtze estuary. Trans Oceanol Limnol 4:35–40
Jiang T, Liu HB, Li MM et al (2018) Investigation on shrimp feeding of Coilia nasus during its anadromous migration along the Yangtze River. J Lake Sci 30(2):458–463
Duan JR, Fang DA, Zhang MY et al (2016) Changes of gonadotropin-releasing hormone receptor 2 during the anadromous spawning migration in Coilia nasus. BMC Dev Biol. https://doi.org/10.1186/s12861-016-0142-9
Lozupone CA, Stombaugh JI, Gordon JI et al (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489(7415):220–230
Wang AR, Ran C, Ring E et al (2018) Progress in fish gastrointestinal microbiota research. Rev Aquac 10:626–640
Nie ZJ, Xu GC, Cheng QQ et al (2014) Construction and analysis of PCR-DGGE gene fingerprint of bacterial community in Coilia nasus from sea. Mar Sci 38(10):68–69
Jiang M, Zhang XZ, Yang YP et al (2019) The effect of Acanthosentis cheni infection on the microbiota composition and diversity in the intestine of the Coilia nasus. J Fish Sci Chin 26(03):176–184
Wang S, Hou X, Su H (2017) Exploration of the relationship between biogas production and microbial community under high salinity conditions. Sci Rep. https://doi.org/10.1038/s41598-017-01298-y
Sullam KE, Essinger SD, Lozupone CA et al (2012) Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol 21:3363–3378
Vermeij GJ, Dudley R (2000) Why are there so few evolutionary transitions between aquatic and terrestrial ecosystems? Biol J Linn Soc 70(4):541–554
Zhang M, Sun Y, Liu Y et al (2016) Response of gut microbiota to salinity change in two euryhaline aquatic animals with reverse salinity preference. Aquaculture 454:72–80
Cahill MM (1990) Bacterial flora of fishes: a review. Microb Ecol 19(1):21–41
Plumb JA (1991) Major diseases of striped bass and redish. Vet Hum toxicol 33(1):34–39
Sonenshein AL, Hoch JA, Losick R (2002) Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington
Sapre S, Gontia-Mishra I, Tiwari S (2018) Klebsiella sp. confers enhanced tolerance to salinity and plant growth promotion in oat seedlings (Avena sativa). Microbiol Res 206:25–32
Kesserü P, Kiss I, Bihari Z et al (2002) The effects of NaCl and some heavy metals on the denitrification activity of Ochrobactrum anthropi. J Basic Microb 42(4):268–276
Muegge BD, Kuczynski J, Dan K et al (2011) Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332(6032):970–974
Margolis L (1953) The effect of fasting on the bacterial flora of the intestine of fish. J Fish Res Board Can 10(2):62–63
Costello EK, Gordon JI, Secor SM et al (2010) Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J 4(11):1375–1385
Löfmark S, Jernberg C, Jansson JK et al (2006) Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. J Antimicrob Chemother 58(6):1160–1167
Crawford PA, Crowley JR, Sambandam N et al (2009) Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci USA 106(27):11276–11281
Vaughan EE, Kleerebezem M, de Vos WM (2003) Genetics of the metabolism of lactose and other sugars. Genet Lactic Acid Bact. https://doi.org/10.1007/978-1-4615-0191-6_4
Ki BM, Kim YM, Jeon JM et al (2017) Characterization of bacterial community dynamics during the decomposition of pig carcasses in simulated soil burial and composting systems. J Microbiol Biotechnol 27(12):2199–2210
Zhang H, Huang T, Liu T (2013) Sediment enzyme activities and microbial community diversity in an oligotrophic drinking water reservoir, eastern China. PLoS ONE. https://doi.org/10.1371/journal.pone.0078571
Zhou C, Song C, Huang D et al (2011) Isolation and characterization of organic phosphorus-mineralizing bacteria in sediment of a Chinese large shallow eutrophic lake (Lake Taihu). Geomicrobiol J 28(8):660–666
Liu L, Zou L, Gao DM (2014) Spatiotemporal distribution of microorganism in mudflats environment of the yellow river estuary intertidal area. Adv Mater Res 1030–1032:603–608
Kohl KD, Amaya J, Passement CA et al (2014) Unique and shared responses of the gut microbiota to prolonged fasting: a comparative study across five classes of vertebrate hosts. FEMS Microbiol Ecol 90(3):883–894
Kiessling A, Lindahl-kiessling K, Kiessling K (2004) Energy utilization and metabolism in spawning migrating Early Smart sock-eye salmon (Oncorhynchus nerka): the migratory paradox. Can J Fish Aquat Sci 61:452–465
Oren A (2001) The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations: implications for the functioning of Salt Lake ecosystems. Hydrobiologia 466(1–3):61–72
Oliphant K, Allen-Vercoe E (2019) Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome. https://doi.org/10.1186/s40168-019-0704-8
Morrison DJ, Preston T (2016) Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7:189–200
Xu G, Du F, Li Y et al (2016) Integrated application of transcriptomics and metabolomics yields insights into population-asynchronous ovary development in Coilia nasus. Sci Rep. https://doi.org/10.1038/srep31835
Acknowledgements
This study was supported by National Key R & D Program of China (2018YFD0900901 & 2019YFD0901203), Major project of hydrobios resources in Jiangsu province (ZYHB16), Species Resources Conservation Project of the Ministry of Agriculture (Investigation and Analysis of Special fishing species Resources), Ecological and Environmental Monitoring system of the three Gorges Project of the Yangtze River (Downstream station) monitoring project (JJ[2017]-010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
There are no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ying, Cp., Jiang, M., You, L. et al. Variations and Potential Factors of Gut Prokaryotic Microbiome During Spawning Migration in Coilia nasus. Curr Microbiol 77, 2802–2812 (2020). https://doi.org/10.1007/s00284-020-02088-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02088-y