Abstract
The function of starch phosphorylase has long been debated on the regulation of starch metabolism during the growth and development of plants. In this study, we isolated starch phosphorylase genes (Pho1 and Pho2) from barley, characterized their gene and protein structures, predicated their promoter’s cis-elements and analyzed expression patterns. Multiple alignments of these genes showed that (1) both Pho1 and Pho2 genes possess 15 exons and 14 introns in all but three of the species analyzed, Aegilops tauschii (for Pho1 which contains 16 exons and 15 introns), potato (for Pho1b which contains 14 exons and 13 introns), and Triticum uraru (for Pho2 which contains 15 exons and 14 introns); (2) the exon–intron junctions of Pho1 and Pho2 flanking the ligand-binding sites are more conservative than the other regions. Analysis of protein sequences revealed that Pho1 and Pho2 were highly homologous except for two regions, the N terminal domain and the L78 insertion region. The results of real-time quantitative PCR (RT-qPCR) indicated that Pho2 is mainly expressed in germinating seeds, and the expression of Pho1 is similar to that of starch synthesis genes during seed development in barley. Microarray-based analysis indicated that the accumulation of Pho1 or Pho2 transcripts exhibited uniform pattern both in various tissues and various stages of seed development among species of barley, rice, and Arabidopsis. Pho1 of barley was significantly down-regulated under cold and drought treatments, and up-regulated under stem rust infection. Pho2 exhibited similar expression to Pho1 in barley. However, significant difference in expression was not detected for either Pho1 or Pho2 under any of the investigated abiotic stresses. In Arabidopsis, significant down-regulation was detected for Pho1 (PHS1) under abscisic acid (ABA) and for Pho2 (PHS2) under cold, salt, and ABA. Our results provide valuable information to genetically manipulate phosphorylase genes and to further elucidate their regulatory mechanism in the starch biosynthetic pathway.






Similar content being viewed by others
Abbreviations
- Aa:
-
Acid residues
- ABA:
-
Abscisic acid
- AGP:
-
Adenosine 5′ diphosphate glucose pyrophosphorylase
- BE:
-
Branch enzyme
- CTAB:
-
Hexadecyltrimethylammonium bromide
- DAF:
-
Days after flowering
- dNTP:
-
Deoxynucleotide triphosphat
- GAPDH:
-
Glycerinaldehyde-phosphate dehydrogenase
- L78:
-
About 80-amino acid long insert region
- NCBI:
-
National Center for Biotechnology Information
- ORF:
-
Open reading frame
- Pho:
-
Phosphorylase
- Pho1 (PHS1):
-
Plastidial phosphorylase
- Pho2 (PHS2):
-
Cytosolic phosphorylase
- PLP:
-
Pyridoxal phosphate
- RT-qPCR:
-
Real-time quantitative polymerase chain reaction
References
Buchner P, Borisjuk L, Wobus U (1996) Glucan phosphorylases in Vicia faba L.: cloning, structural analysis and expression patterns of cytosolic and plastidic forms in relation to starch. Planta 199:64–73
Dauvillee D, Chochois V, Steup M, Haebel S, Eckermann N, Ritte G, Ral JP, Colleoni C, Hicks G, Wattebled F, Deschamps P, d’Hulst C, Lienard L, Cournac L, Putaux JL, Dupeyre D, Ball SG (2006) Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J 48:274–285
Duwenig E, Steup M, Kossmann J (1997) Induction of genes encoding plastidic phosphorylase from spinach (Spinacia oleracea L.) and potato (Solanum tuberosum L.) by exogenously supplied carbohydrates in excised leaf discs. Planta 203:111–120
Higgins JE, Kosar-Hashemi B, Li Z, Howitt CA, Larroque O, Flanagan B, Morell MK, Rahman S (2013) Characterization of starch phosphorylases in barley grains. J Sci Food Agric 93:2137–2145
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Hudson JW, Golding GB, Crerar MM (1993) Evolution of allosteric control in glycogen phosphorylase. J Mol Biol 234:700–721
Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, De Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ (2008) The 20 years of PROSITE. Nucleic Acids Res 36:245–249
James MG, Denyer K, Myers AM (2003) Starch synthesis in the cereal endosperm. Curr Opin Plant Biol 6:215–222
Jiang QT, Liu T, Ma J, Wei YM, Lu ZX, Lan XJ, Dai SF, Zheng YL (2011) Characterization of barley Prp1 gene and its expression during seed development and under abiotic stress. Genetica 139:1283–1292
Keren H, Lev-Maor G, Ast G (2010) Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11:345–355
Lin YC, Chen HM, Chou IM, Chen AN, Chen CP, Young GH, Lin CT, Cheng CH, Chang SC, Juang RH (2012) Plastidial starch phosphorylase in sweet potato roots is proteolytically modified by protein–protein interaction with the 20S proteasome. PLoS One 7(4):e35336
Long XY, Wang JR, Ouellet T, Rocheleau H, Wei YM, Pu Z, Jiang QT, Lan XJ, Zheng YL (2010) Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol Biol 74:307–311
Ma J, Jiang QT, Zhao QZ, Zhao S, Lan XJ, Dai SF, Lu ZX, Liu CJ, Wei YM, Zheng YL (2013) Characterization and expression analysis of waxy alleles in barley accessions. Genetica 141:227–238
Mori H, Tanizawa K, Fukui T (1993) A chimeric alpha-glucan phosphorylase of plant type L and H isozymes. Functional role of 78-residue insertion in type L isozyme. J Biol Chem 268:5574–5581
Mu HH, Yu Y, Wasserman BP, Carman GM (2001) Purification and characterization of the maize amyloplast stromal 112-kDa starch phosphorylase. Arch Biochem Biophys 388:155–164
Murray M, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Nakamura Y, Ono M, Utsumi C, Steup M (2012) Functional interaction between plastidial starch phosphorylase and starch branching enzymes from rice during the synthesis of branched maltodextrins. Plant Cell Physiol 53:869–878
Nakano K, Fukui T (1986) The complete amino acid sequence of potato alpha-glucan phosphorylase. J Biol Chem 261:8230–8236
Newgard CB, Hwang PK, Fletterick RJ (1989) The family of glycogen phosphorylases: structure and function. Crit Rev Biochem Mol Biol 24:69–99
Preiss J, Levi C (1980) Starch biosynthesis and degradation. In: Preiss J (ed) The biochemistry of plants, vol 3. Academic Press, London, pp 371–423
Radchuk VV, Borisjuk L, Sreenivasulu N, Merx K, Mock HP, Rolletschek H, Wobus U, Weschke W (2009) Spatiotemporal profiling of starch biosynthesis and degradation in the developing barley grain. Plant Physol 150:190–204
Rathore R, Garg N, Garg S, Kumar A (2009) Starch phosphorylase: role in starch metabolism and biotechnological applications. Crit Rev Biotechnol 29:214–224
Reddy AS (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58:267–294
Regina A, Blazek J, Gilbert E, Flanagan BM, Gidley MJ, Cavanagh C, Ral J-P, Larroque O, Bird AR, Li Z, Morell MK (2012) Differential effects of genetically distinct mechanisms of elevating amylose on barley starch characteristics. Carbohyd Polym 89:979–991
Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X (2009) DOG 1.0: illustrator of protein domain structures. Cell Res 19:271–273
Satoh H, Shibahara K, Tokunaga T, Nishi A, Tasaki M, Hwang S-K, Okita TW, Kaneko N, Fujita N, Yoshida M (2008) Mutation of the plastidial α-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20:1833–1849
Schupp N, Ziegler P (2004) The relation of starch phosphorylases to starch metabolism in wheat. Plant Cell Physiol 45:1471–1484
Sonnewald U, Basner A, Greve B, Steup M (1995) A second L-type isozyme of potato glucan phosphorylase: cloning, antisense inhibition and expression analysis. Plant Mol Biol 27:567–576
Steup M, Robenek H, Melkonian M (1983) In-vitro degradation of starch granules isolated from spinach chloroplasts. Planta 158:428–436
Streb S, Eicke S, Zeeman SC (2012) The simultaneous abolition of three starch hydrolases blocks transient starch breakdown in Arabidopsis. J Biol Chem 287: 41745–41756
Subasinghe RM (2013) Role and regulation of starch phosphorylase and starch synthase IV in starch biosynthesis in maize endosperm amyloplasts. Dissertation. The University of Guelph, Canada
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tetlow IJ (2011) Starch biosynthesis in developing seeds. Seed Sci Res 21:5
Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emes MJ (2004) Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein–protein interactions. Plant Cell 16:694–708
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tickle P, Burrell MM, Coates SA, Emes MJ, Tetlow IJ, Bowsher CG (2009) Characterization of plastidial starch phosphorylase in Triticum aestivum L. endosperm. J Plant Physiol 166:1465–1478
Tiwari R, Kumar A (2012) Starch phosphorylase: biochemical and biotechnological perspectives. Biotechnol Mol Biol Rev 7:69–83
Van Hung P, Maeda T, Morita N (2006) Waxy and high-amylose wheat starches and flours -Characteristics, functionality and application. Trends Food Sci Technol 17:448–456
Weschke W, Panitz R, Sauer N, Wang Q, Neubohn B, Weber H, Wobus U (2000) Sucrose transport into barley seeds: molecular characterization of two transporters and implications for seed development and starch accumulation. Plant J 21:455–467
Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480:520–524
Yu Y, Mu HH, Wasserman BP, Carman GM (2001) Identification of the maize amyloplast stromal 112-kD protein as a plastidic starch phosphorylase. Plant Physiol 125:351–359
Zeeman SC, Thorneycroft D, Schupp N, Chapple A, Weck M, Dunstan H, Haldimann P, Bechtold N, Smith AM, Smith SM (2004) Plastidial alpha-glucan phosphorylase is not required for starch degradation in Arabidopsis leaves but has a role in the tolerance of abiotic stress. Plant Physiol 135:849–858
Zeeman SC, Kossmann J, Smith AM (2010) Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 61:209–234
Zouine M, Latché A, Rousseau C, Regad F, Pech J-C, Philippot M, Bouzayen M, Delalande C, Frasse P, Schiex T (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31000167 and 31230053) and the China Transgenic Research Program (2011ZX08002-001,004 and 005). We appreciate the referees for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Jian Ma and Qian-Tao Jiang authors contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2013_1953_MOESM2_ESM.tif
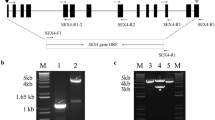
Supplementary Fig. S1 Structure and isolation of Pho1 and Pho2 genes. a Aegilops tauschii Pho1 gene structure and PCR primers for amplicaton of barley Pho1. b Aegilops tauschii Pho2 gene structure and PCR primers locations for barley Pho2. c PCR banding pattern of barley Pho1 and Pho2 genes. Boxes and thick lines present exons and introns, respectively. The positions of primers (see Table 1) are indicated by arrows. Triangles illustrate the approximate positons of the start of (ATG) and the stop (TGA) codons. 1, Pho1-F2/R2; 2, Pho1-F3/R3; 3, Pho1-F4/R4; 4, Pho1-F5/R5; 5, Pho1-F6/R6; 6, Pho2-F1v/R1v; 7, Pho2-F2/R2; 8, Pho2-F3/R3; 9, Pho2-F4/R4; 10, Pho2-F5/R5; 11, Pho2-F1v2/R3; 12, Pho2-F4/R5. M. DNA 1kb plus ladder (TIFF 3328 kb)
425_2013_1953_MOESM3_ESM.tif
Supplementary Fig. S2 Multiple alignments of Pho1 and Pho2 amino acid sequences among different species using CLC Main Workbench 6.7.2. Hv, barley; Ta, Triticum aestivum; At, Arabidopsis thaliana; St, Solanum tuberosum; Cr, Chlamydomonas reinhardtii; Ca, Caldilinea aerophila; Hs, Homo sapiens (muscle) (TIFF 192 kb)
Rights and permissions
About this article
Cite this article
Ma, J., Jiang, QT., Zhang, XW. et al. Structure and expression of barley starch phosphorylase genes. Planta 238, 1081–1093 (2013). https://doi.org/10.1007/s00425-013-1953-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-013-1953-6