Abstract
Main conclusion
Comparative morphological, transcriptomic and phytohormone analyses reveal a defence network leading to PCD involved in cabbage hybrid lethality.
Abstract
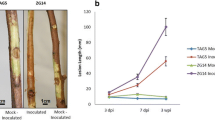
Hybrid lethality (HL) plays an essential role in the stability of a population by blocking gene exchange between species, but the molecular mechanism remains largely undetermined. In this study, we performed phenotype, transcriptome and plant hormone analyses of HL in cabbage. Phenotype analysis confirmed that HL is characterised by a typical programmed cell death (PCD) process. A time-resolved RNA-Seq identified 2724 differentially expressed genes (DEGs), and functional annotations analyses revealed that HL was closely associated with the defence response. A defence regulation network was constructed based on the plant–pathogen interaction pathway and MAPK signalling pathway, which comprised DEGs related to Ca2+ and hydrogen peroxide (H2O2) leading to PCD. Moreover, important DEGs involved in hormone signal transduction pathways including salicylic acid (SA) and jasmonic acid (JA) were identified, which were further confirmed by endogenous and exogenous SA and JA measurements. Our results identified key genes and pathways in the regulating network of HL in cabbage, and might open the gate for revealing the molecular mechanism of HL in plants.










Similar content being viewed by others
Abbreviations
- DEG:
-
Differentially expressed gene
- HL:
-
Hybrid lethality
- HR:
-
Hypersensitive response
- JA:
-
Jasmonic acid
- PCD:
-
Programmed cell death
- SA:
-
Salicylic acid
- TF:
-
Transcription factor
References
Aleo M (2002) Endoplasmic reticulum Ca2+ signaling and calpains mediate renal cell death. Cell Death Differ 9:734–741 https://doi.org/10.1038/sj.cdd.44010291
Alcazar R, Garcia AV, Parker JE, Reymond M (2009) Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. P Natl Acad Sci 106:334–339. https://doi.org/10.1073/pnas.0811734106
Antoniw J, Ritter C, Pierpoint W, Van Loon L (1980) Comparison of three pathogenesis-related proteins from plants of two cultivars of tobacco infected with TMV. J Gen Virol 47:79–87. https://doi.org/10.1099/0022-1317-47-1-79
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant 55:373–399. https://doi.org/10.1146/annurev.arplant.55.031903.141701
Bari R, Jones J (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69:473–488. https://doi.org/10.1007/s11103-008-9435-0
Beckers G, Spoel S (2006) Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biol 8:1–10. https://doi.org/10.1055/s-2005-872705
Bomblies KJ, Lempe P, Epple N, Warthmann C, Lanz J, Dangl L, Weigel D (2007) Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol 5:1962–1972. https://doi.org/10.1371/journal.pbio.0050236
Chae E, Bomblie K, Kim S, Karelin D, Zaidem M, Ossowski S, Martín-Pizarro C, Laitinen R, Rowan B, Tenenboim H, Lechner S, Demar M, Habring-Müller A, Lanz C, Rätsch G, Weigel D (2014) Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159:1341–1351. https://doi.org/10.1016/j.cell.2014.10.049
Chen C, Chen H, Shan J, Zhu M, Shi M, Gao J, Lin H (2013) Genetic and physiological analysis of a novel type of interspecific hybrid weakness in rice. Mol Plant 6:716–728. https://doi.org/10.1093/mp/sss146
Chini A, Fonseca S, Fernández G, Adie B, Chico J, Lorenzo O, García-Casado G, López-Vidriero I, Lozano F, Ponce M, Micol J, Solano R (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448:666–671. https://doi.org/10.1038/nature06006
Cho D, Shin D, Jeon B, Kwak J (2009) ROS-mediated ABA signaling. J Plant Biol 52:102–113. https://doi.org/10.1007/s12374-009-9019-9
Clay N, Adio A, Carine D, Georg J, Ausubel F (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323:95–101. https://doi.org/10.1126/science.1164627
Deng J, Fang L, Zhu X, Zhou B, Zhang T (2019) CC-NBS-LRR gene induces hybrid lethality in cotton. J Exp Bot 70:5145–5156. https://doi.org/10.1093/jxb/erz312
Dhirendra K (2014) Salicylic acid signaling in disease resistance. Plant Sci 228:127–134. https://doi.org/10.1016/j.plantsci.2014.04.014
Dobzhansky T (1959) Genetics and the origin of species. Nature 184:587–588. https://doi.org/10.1093/aibsbulletin/2.2.14-b
Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE (1999) EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA 96:3292–3297. https://doi.org/10.1073/pnas.96.6.3292
Flor H (2003) Current status of the gene-for-gene concept. Annu Rev Phytopathol 9:275–296. https://doi.org/10.1146/annurev.py.09.090171.001423
Häffner E, Karlovsky P, Splivallo R, Traczewska A, Diederichsen E (2014) ERECTA, salicylic acid, abscisic acid, and jasmonic acid modulate quantitative disease resistance of Arabidopsis thaliana to Verticillium longisporum. BMC Plant Biol 14:85. https://doi.org/10.1186/1471-2229-14-85
Hannah M, Krmer K, Geffroy V, Kopka BM, Erban A, Vallejos C, Heyer A, Sanders F, Millneret P (2007) Hybrid weakness controlled by the dosage-dependent lethal (DL) gene system in common bean (Phaseolus vulgaris) is caused by a shoot-derived inhibitory signal leading to salicylic acid-associated root death. New Phytol 176:537–549. https://doi.org/10.1111/j.1469-8137.2007.02215.x
Hollingshead L (1929) Lethal factor in Crepis effective only in an interspecific hybrid. Genetics 15:114–140. https://doi.org/10.1007/BF02983371
Hu Y, Xue Y, Liu J, Fang Z, Yang L, Zhang Y, Lv H, Liu Y, Li Z, Zhuang M (2016) Hybrid lethality caused by two complementary dominant genes in cabbage (Brassica oleracea L). Mol Breed 36:73. https://doi.org/10.1007/s11032-016-0498-3
Imran H, Zhang Y, Du G (2007) Effect of salicylic acid (SA) on delaying fruit senescence of Huang Kum pear. Front Agric China 1:456–459. https://doi.org/10.1007/s11703-007-0075-y
Jeuken MJ, Zhang NW, McHale LK, Pelgrom KE, Lindhout B, Michelmore R, Vissera R, Niksa R (2009) Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21:3368–3378. https://doi.org/10.1105/tpc.109.070334
Kjaersgaard T, Jensen M, Christiansen M, Gregersen P, Skriver K (2011) Senescence-associated barley NAC (NAM, ATAF1, 2, CUC) transcription factor interacts with radical-induced cell death 1 through a disordered regulatory domain. J Biol Chem 286:35418–35429. https://doi.org/10.1074/jbc.M111.247221
Koo A, Gao X, Jones A, Howe G (2009) A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J 59:974–986. https://doi.org/10.1111/j.1365-313X.2009.03924.x
Kunkel B (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant. https://doi.org/10.1016/S1369-5266(02)00275-3
Livak K, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402. https://doi.org/10.1006/meth.2001.1262
Maheshwari S, Barbash D (2010) The genetics of hybrid incompatibilities. Annu Rev Genet 45:331–355. https://doi.org/10.1146/annurev-genet-110410-132514
Mitsuhara I, Takayoshi I, Shigemi S, Yuki Y, Hiroyuki K, Sakino H (2008) Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol Genet Genomics 279:415–427. https://doi.org/10.1007/s00438-008-0322-9
Mittler R, Vanderauwera S, Gollery M, Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498. https://doi.org/10.1016/j.tplants.2004.08.009
Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113:935–944. https://doi.org/10.1016/S0092-8674(03)00429-X
Muller HJ (1942) Isolating mechanisms, evolution and temperature. Biol Symp 6:71–125
Orr HA, Presgraves DC (2000) Speciation by postzygotic isolation: forces, genes and molecules. BioEssays 22:1085–1094. https://doi.org/10.1002/1521-1878(200012)22:12%3c1085::AID-BIES6%3e3.0.CO;2-G
Pauwels P, Josée E, Janssen P (1991) Ca++ and Na+ channels involved in neuronal cell death protection by flunarizine. Life Sci 48:1881–1893. https://doi.org/10.1016/0024-3205(91)90220-6
Pickersgill B (1971) Relationships between weedy and cultivated forms in some species of Chili peppers (genus Capsicum). Evolution 25:683–691. https://doi.org/10.2307/2406949
Pre M, Atallah M, Champion A, De Vos M, Pieterse C, Memelink J (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147:1347–1357. https://doi.org/10.1104/pp.108.117523
Rubén A, Ana V, Jane E, Matthieu R (2009) Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci USA 106:334–339. https://doi.org/10.1073/pnas.0811734106
Shii C, Mok T, Temple S, Mok D (1980) Expression of developmental abnormalities in hybrids of Phaseolus vulgaris L. Interaction between temperature and allelic dosage. J Hered 71:219–222. https://doi.org/10.1093/oxfordjournals.jhered.a109353
Straus MR, Rietz S, Ver L, Themaat E, Bartsch M, Parker J (2010) Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J 62:628–640. https://doi.org/10.1111/j.1365-313X.2010.04178.x
Tang D (2005) Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in Arabidopsis by the EDR1 protein kinase. Plant Physiol 138:1018–1026. https://doi.org/10.1104/pp.105.060400
Tezuka T, Marubashi W (2006) Hybrid lethality in interspecific hybrids between Nicotiana tabacum and N. suaveolens: evidence that the Q chromosome causes hybrid lethality based on Q-chromosome-specific DNA markers. Theor Appl Genet 112:1172–1178. https://doi.org/10.1007/s00122-006-0219-0
Ting Z, Xu P, Wang W, Wang S, Caruana J, Yang H (2018) Arabidopsis g-protein β subunit AGB1 interacts with BES1 to regulate brassinosteroid signaling and cell elongation. Front Plant Sci 8:2225. https://doi.org/10.3389/fpls.2017.02225
Tomoya N, Ichiro Shigemi S, Norihiro O, Yuko O (1998) Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol 5:1662–1670. https://doi.org/10.1086/498022
Verbert L, Devogelaere B, Parys J, Missiaen L, de Smedt H (2007) Proteolytic mechanisms leading to disturbed Ca2+ signaling in apoptotic cell death. Calcium Bind Proteins 2(1):21–29
Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker J (2013) Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14:619–630. https://doi.org/10.1016/j.chom.2013.11.006
Wang Y, Hao X, Lu Q, Wang L, Qian W, Li N, Ding C, Wang X, Yang Y (2018) Transcriptional analysis and histochemistry reveal that hypersensitive cell death and H2O2 have crucial roles in the resistance of tea plant (Camellia sinensis (L.) O. Kuntze) to anthracnose. Hortic Res 1:18. https://doi.org/10.1038/s41438-018-0025-2
Wiebe G (1934) Complementary factors in barley giving a lethal progeny. J Hered 25:273–274. https://doi.org/10.1093/oxfordjournals.jhered.a103943
Xiao Z, Hu Y, Zhang X, Xue Y, Fang Z, Yang L, Zhang Y, Liu Y, Li Z, Liu X, Liu Z, Zhuang M (2017) Fine mapping and transcriptome analysis reveal candidate genes associated with hybrid lethality in cabbage (Brassica oleracea). Genes 5:8. https://doi.org/10.3390/genes8060147
Xing M, Su H, Liu X, Yang L, Zhang Y, Wang Y, Fang Z, Lv H (2019) Morphological, transcriptomics and phytohormone analysis shed light on the development of a novel dwarf mutant of cabbage (Brassica oleracea). Plant Sci 290:110283. https://doi.org/10.1016/j.plantsci.2019.110283
Yamada T, Marubashi W (2003) Overproduced ethylene causes programmed cell death leading to temperature-sensitive lethality in hybrid seedlings from the cross Nicotiana suaveolens × N. tabacum. Planta 217:690–698. https://doi.org/10.1007/s00425-003-1035-2
Yamauchi T, Yoshioka M, Fukazawa A, Mori H, Nishizawa N, Tsutsumi N, Yoshioka H, Nakazono M (2017) An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 29:775–790. https://doi.org/10.1105/tpc.16.00976
Yang C, Li W, Cao J, Meng F, Liu J (2016) Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J 89:338. https://doi.org/10.1111/tpj.13388
Zhou W, Yoshida K, Shintaku Y, Takeda G (1991) The use of IAA to overcome interspecific hybrid inviability in reciprocal crosses between Nicotiana tabacum L. and N. repanda Willd. Theor Appl Genet 82:657–661. https://doi.org/10.1007/BF00226805
Zuellig M, Sweigart A, Malik H (2018) Gene duplicates cause hybrid lethality between sympatric species of Mimulus. PLoS Genet 14:e1007130. https://doi.org/10.1371/journal.pgen.1007130
Acknowledgements
The work reported here was performed at the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, Beijing 100081, China.
Funding
This work was financially supported by grants from the National Natural Science Foundation of China (31572139), the Key Projects of the National Key Research and Development Program of China (2016YFD0100307), the Central Public-interest Scientific Institution Basal Research Fund (Y2020PT01), and the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiao, Z., Liu, X., Fang, Z. et al. Transcriptome and plant hormone analyses provide new insight into the molecular regulatory networks underlying hybrid lethality in cabbage (Brassica oleracea). Planta 253, 96 (2021). https://doi.org/10.1007/s00425-021-03608-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-021-03608-1