Abstract
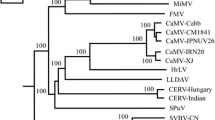
The complete sequence of a strawberry vein banding virus (SVBV) isolate collected in Nova Scotia, Canada, and designated NS8, was determined. The 7,856-nucleotide circular double-stranded DNA genome contains seven open-reading frames (ORFs), which is consistent with other SVBV isolates and other members of the genus Caulimovirus. Comparison of NS8 with other whole-genome sequences retrieved from databases revealed that NS8 shares the highest sequence similarity (96.5% identity) with isolate China (accession number HE681085) and the lowest (88.3% identity) with clone pSVBV-E3 (accession number X97304). Despite the overall high sequence similarity between NS8 and China, the coat protein encoding ORF IV of NS8 shares only 90.9% sequence identity with the China isolate. Phylogenetic analysis at the complete-genome level placed NS8 and all Chinese isolates in one clade and clone pSVBV-E3 in a separate clade. Interestingly, phylogenetic analysis of all available ORF IV sequences, including those retrieved from databases and newly sequenced samples in this study from Canada, revealed three distinct clades. All Canadian isolates grouped together as one clade, pSVBV-E3 and several others from Europe, Egypt and the USA grouped as a second clade, and isolates from China formed a third clade. These results demonstrate that SVBV is more divergent than previously reported.

Similar content being viewed by others
References
Anonymous (2014) Statistical Overview of the Canadian Fruit Industry—2014. http://www.agr.gc.ca/eng/industry-markets-and-trade/statistics-and-market-information/by-product-sector/horticulture/horticulture-canadian-industry/sector-reports/statistical-overview-of-the-canadian-fruit-industry-2014/?id=1447784197130
Bhagwat B, Dickison V, Su L, Bernardy M, Wiersma PA, Nie X, Xiang Y (2016) Molecular characterization of divergent strawberry mild yellow edge virus isolates from eastern Canada. J Phytopathol 164:691–696
Bhagwat B, Dickison V, Ding X, Walker M, Bernardy M, Bouthillier M, Creelman A, DeYoung R, Li Y, Nie X, Wang A, Xiang Y, Sanfaçon H (2016) Genome sequence analysis of five Canadian isolates of strawberry mottle virus reveals extensive intra-species diversity and a longer RNA2 with increased coding capacity compared to a previously characterized European isolate. Arch Virol 161:1657–1663
Chang L, Zhang Z, Yang H, Li H, Dai H (2007) Detection of strawberry RNA and DNA viruses by RT-PCR using total nucleic acid as a template. J Phytopathol 155:431–436
Chenault KD, Melcher U (1994) Phylogenetic relationships reveal recombination among isolates of cauliflower mosaic virus. J Mol Evol 39:496–505
Ding X, Li Y, Hernández-Sebastià C, Abbasi PA, Fisher P, Celetti MJ, Wang A (2016) First report of Strawberry crinivirus 4 on strawberry in Canada. Plant Dis 100:1254
Farzadfar S, Pourrahim R, Ebrahimi H (2014) A phylogeographical study of the cauliflower mosaic virus population in mid-Eurasia Iran using complete genome analysis. Arch Virol 159:1329–1340
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol. doi:10.1093/molbev/msw054
Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160
Ma X, Cui H, Bernardy M, Tian L, Abbasi Wang A (2015) Molecular characterization of a Strawberry mild yellow edge virus isolate from Canada. Can J Plant Pathol 37:369–375
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003
Martin RR, Tzanetakis IE (2013) High risk strawberry viruses by region in the United States and Canada: implications for certification, nurseries, and fruit production. Plant Dis 97:1358–1362
Mráz I, Petrzik K, Šíp M, Fránová-Honetšlegrová J (1998) Variability in coat protein sequence homology among American and European sources of strawberry vein banding virus. Plant Dis 82:544–546
Ng JCK, Perry KL (2004) Transmission of plant viruses by aphid vectors. Mol Plant Pathol 5:505–511
Pattanaik S, Dey N, Bhattacharyya S, Maiti IB (2004) Isolation of full-length transcript promoter from the Strawberry vein banding virus (SVBV) and expression analysis by protoplasts transient assays and in transgenic plants. Plant Sci 167:427–438
Petrzik K, Beneš V, Mráz I, Honetšlegrová-Fránová J, Ansorge W, Špak J (1998) Strawberry vein banding virus - definitive member of the genus Caulimovirus. Virus Genes 16:303–305
Ragab M, El-Dougdoug K, Mousa S, Attia A, Sobolev I, Spiegel S, Freeman S, Zeidan M, Tzanetakis IE, Martin RR (2009) Detection of strawberry viruses in Egypt. Acta Hort 842:319–322
Ratti C, Pisi A, Rubies Autonell C, Babini A, Vicchi V (2009) First report of Strawberry vein banding virus on strawberry in Italy. Plant Dis 93:675
Sofy AR (2015) First molecular characterization of Strawberry vein banding virus naturally spread in mixed infection with fruit phyllody phytoplasma in Egypt. Int J Virol Mol Biol 4:23–36
Stenger DC, Mullin RH, Morris TJ (1988) Isolation, molecular cloning, and detection of strawberry vein banding virus DNA. Mol Plant Pathol 78:154–159
Thompson JR, Wetzel S, Klerks MM, Vašková D, Schoen CD, Špak J, Jelkmann W (2003) Multiplex RT-PCR detection of four aphid-borne strawberry viruses in Fragaria spp. in combination with a plant mRNA specific internal control. J Virol Methods 111:85–93
Tzanetakis IE, Martin RR (2013) Expanding field of strawberry viruses which are important in North America. Int J Fruit Sci 13:184–195
Xiang Y, Bernardy M, Bhagwat B, Wiersma PA, DeYoung R, Bouthillier M (2015) The complete genome sequence of a new polerovirus in strawberry plants from eastern Canada showing strawberry decline symptoms. Arch virol 160:553–556
Yoshikawa N, Inouye T (1988) Strawberry viruses occurring in Japan. Acta Hort 236:59–67
Acknowledgements
We thank Dr. Pervaiz Abbasi for providing strawberry plants, and Darcy Sutherland for technical assistance. We also acknowledge the participation and contribution of Dylan Woodcock at the early stage of this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was funded by Agriculture and Agri-Food Canada under the project J-000051. All authors declare that they have no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2017_3252_MOESM2_ESM.jpg
Supplementary Figure 1 Analysis of sequence identities between the query sequence (A. pSVBV-E3; B. BJ; C. Shenyang; D. China) and other isolates (reference sequences) using SimPlot [9]. Similarities (y-axis, 0.5-1) are plotted along the nucleotide sequence (x-axis, nt 1-ca. 7,800). The window covered 200 nucleotides and moved along the alignment in 20-nucleotide steps (JPEG 2843 kb)
Rights and permissions
About this article
Cite this article
Dickison, V., MacKenzie, T.D.B., Singh, M. et al. Strawberry vein banding virus isolates in eastern Canada are molecularly divergent from other isolates. Arch Virol 162, 1777–1781 (2017). https://doi.org/10.1007/s00705-017-3252-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3252-1