Abstract
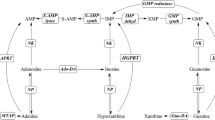
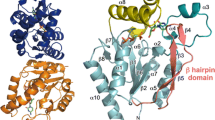
The degradation of purine nucleoside is the first step of purine nucleoside uptake. This degradation is catalyzed by purine nucleoside phosphorylase, which is categorized into two classes: hexameric purine nucleoside phosphorylase (6PNP) and trimeric purine nucleoside phosphorylase (3PNP). Generally, 6PNP and 3PNP degrade adenosine and guanosine, respectively. However, the substrate specificity of 6PNP and 3PNP of Thermus thermophilus (tt6PNP and tt3PNP, respectively) is the reverse of that anticipated based on comparison to other phosphorylases. Specifically, in this paper we reveal by gene disruption that tt6PNP and tt3PNP are discrete enzymes responsible for the degradation of guanosine and adenosine, respectively, in T. thermophilus HB8 cells. Sequence comparison combined with structural information suggested that Asn204 in tt6PNP and Ala196/Asp238 in tt3PNP are key residues for defining their substrate specificity. Replacement of Asn204 in tt6PNP with Asp changed the substrate specificity of tt6PNP to that of a general 6PNP. Similarly, substitution of Ala196 by Glu and Asp238 by Asn changed the substrate specificity of tt3PNP to that of a general 3PNP. Our results indicate that the residues at these positions determine substrate specificity of PNPs in general. Sequence analysis further suggested most 6PNP and 3PNP enzymes in thermophilic species belonging to the Deinococcus-Thermus phylum share the same critical residues as tt6PNP and tt3PNP, respectively.





Similar content being viewed by others
Abbreviations
- NMP:
-
Nucleoside monophosphate
- PNP:
-
Purine nucleoside phosphorylase
- 6PNP:
-
Hexameric purine nucleoside phosphorylase
- 3PNP:
-
Trimetric purine nucleoside phosphorylase
- tt6PNP:
-
6PNP of T. thermophilus HB8
- tt3PNP:
-
3PNP of T. thermophilus HB8
- Δtt6PNP :
-
tt6PNP gene null mutant
- Δtt3PNP :
-
tt3PNP gene null mutant
- WT:
-
Wild type
- Cv:
-
Column volume(s)
References
Afshar S, Sawaya MR, Morrison SL (2009) Structure of a mutant human purine nucleoside phosphorylase with the prodrug, 2-fluoro-2′-deoxyadenosine and the cytotoxic drug, 2-fluoroadenine. Protein Sci 18:1107–1114
Almendros M, Berenguer J, Sinisterra J-V (2012) Nucleoside phosphorylases from Thermus thermophilus active in the synthesis of nucleoside analogues. Appl Environ Microbiol 78:3128–3135
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bennett EM, Anand R, Allan PW, Hassan AE, Hong JS, Levasseur DN, McPherson DT, Parker WB, Secrist JA III, Sorscher EJ, Townes TM, Waud WR, Ealick SE (2003) Designer gene therapy using an Escherichia coli purine nucleoside phosphorylase/prodrug system. Chem Biol 10:1173–1181
Bzowska A, Kulikowska E, Shugar D (2000) Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol Ther 88:349–425
Canduri F, Silva RG, dos Santos DM, Palma MS, Basso LA, Santos DS, de Azevedo WF Jr (2005) Structure of human PNP complexed with ligands. Acta Crystallogr D Biol Crystallogr 61:856–862
Dessanti P, Zhang Y, Allegrini S, Tozzi MG, Sgarrella F, Ealick SE (2012) Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase. Acta Crystallogr Sect D: Biol Crystallogr 68:239–248
Ducati RG, Santos DS, Basso LA (2009) Substrate specificity and kinetic mechanism of purine nucleoside phosphorylase from Mycobacterium tuberculosis. Arch Biochem Biophys 486:155–164
Erion MD, Stoeckler JD, Guida WC, Walter RL, Ealick SE (1997a) Purine nucleoside phosphorylase. 2. Catalytic mechanism. Biochemistry 36:11735–11748
Erion MD, Takabayashi K, Smith HB, Kessi J, Wagner S, Hönger S, Shames SL, Ealick SE (1997b) Purine nucleoside phosphorylase. 1. Structure-function studies. Biochemistry 36:11725–11734
Giblett ER, Ammann AJ, Wara DW, Sandman R, Diamond LK (1975) Nucleoside-phosphorylase deficiency in a child with severely defective T-cell immunity and normal B-cell immunity. Lancet 1:1010–1014
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Hashimoto Y, Yano T, Kuramitsu S, Kagamiyama H (2001) Disruption of Thermus thermophilus genes by homologous recombination using a thermostable kanamycin-resistant marker. FEBS Lett 506:231–234
Hoseki J, Yano T, Koyama Y, Kuramitsu S, Kagamiyama H (1999) Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for Thermus thermophilus. J Biochem (Tokyo) 126:951–956
Iwai T, Kuramitsu S, Masui R (2004) The Nudix hydrolase Ndx1 from Thermus thermophilus HB8 is a diadenosine hexaphosphate hydrolase with a novel activity. J Biol Chem 279:21732–21739
Jensen KF (1978) Two purine nucleoside phosphorylases in Bacillus subtilis. Purification and some properties of the adenosine-specific phosphorylase. Biochim Biophys Acta 525:346–356
Jensen K, Nygaard P (1975) Purine nucleoside phosphorylase from Escherichia coli and Salmonella typhimurium. Purification and some properties. Eur J Biochem 51:253–265
Kalckar HM (1947a) Differential spectrophotometry of purine compounds by means of specific enzymes; determination of adenine compounds. J Biol Chem 167:445–459
Kalckar HM (1947b) Differential spectrophotometry of purine compounds by means of specific enzymes; determination of hydroxypurine compounds. J Biol Chem 167:429–443
Kalckar HM (1947c) Differential spectrophotometry of purine compounds by means of specific enzymes; studies of the enzymes of purine metabolism. J Biol Chem 167:461–475
Kilstrup M, Hammer K, Ruhdal Jensen P, Martinussen J (2005) Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol Rev 29:555–590
Kim DW, Choi SH, Kang A, Nam SH, Kim DS, Kim RN, Kim A, Park HS (2011) Draft genome sequence of Lactobacillus malefermentans KCTC 3548. J Bacteriol 193:5537
Kleywegt GJ (2007) Crystallographic refinement of ligand complexes. Acta Crys D Biol Crystallogr 63:94–100
Kuramitsu S, Hiromi K, Hayashi H, Morino Y, Kagamiyama H (1990) Pre-steady state kinetics of Escherichia coli aspartate aminotransferase catalyzed reactions and thermodynamic aspects of its substrate specificity. Biochemistry 29:5469–5476
Liacouras AS, Garvey TQ III, Millar FK, Anderson EP (1975) Uridine cytidine kinase: kinetic studies and reaction mechanism. Arch Biochem Biophys 168:74–80
Mikleušević G, Štefanić Z, Narczyk M, Wielgus-Kutrowska B, Bzowska A, Luić M (2011) Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies. Biochimie 93:1610–1622
Neuhard J, Kelln RA (1996) Biosynthesis and conversions of pyrimidines. In: Neidhardt FC (ed) Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol 1. American Society for Microbiology, Washington, DC, pp 1–20
Parks RE Jr, Agarwal RP (1972) Elimination and addition, aldol cleavage and condensation, other C–C cleavage, phosphorolysis, hydrolysis (fats, glycosides). In: Boyer PD (ed) Enzymes, vol 7. Academic Press, San Diego, pp 483–514
Pereira HM, Rezende MM, Castilho MS, Oliva G, Garratt RC (2010) Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni. Acta Crys D Biol Crystallogr 66:73–79
Postigo MP, Guido RVC, Oliva G, Castilho MS, da R. Pitta I, de Albuquerque JFC, Andricopulo AD (2010) Discovery of new inhibitors of Schistosoma mansoni PNP by pharmacophore-based virtual screening. J Chem Inf Model 50:1693–1705
Pukall R, Zeytun A, Lucas S, Lapidus A, Hammon N, Deshpande S, Nolan M, Cheng JF, Pitluck S, Liolios K, Pagani I, Mikhailova N, Ivanova N, Mavromatis K, Pati A, Tapia R, Han C, Goodwin L, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Brambilla EM, Rohde M, Göker M, Detter JC, Woyke T, Bristow J, Eisen J, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP (2011) Complete genome sequence of Deinococcus maricopensis type strain (LB-34). Stand Genomic Sci 4:163–172
Robertson BC, Hoffee PA (1973) Purification and properties of purine nucleoside phosphorylase from Salmonella typhimurium. J Biol Chem 248:2040–2043
Sakon H, Nagai F, Morotomi M, Tanaka R (2008) Sutterella parvirubra sp. nov. and Megamonas funiformis sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 58:970–975
Stoeckler JD, Poirot AF, Smith RM, Parks RE Jr, Ealick SE, Takabayashi K, Erion MD (1997) Purine nucleoside phosphorylase. 3. Reversal of purine base specificity by site-directed mutagenesis. Biochemistry 36:11749–11756
Tahirov T, Inagaki E, Ohshima N, Kitao T, Kuroishi C, Ukita Y, Takio K, Kobayashi M, Kuramitsu S, Yokoyama S, Miyano M (2004) Crystal structure of purine nucleoside phosphorylase from Thermus thermophilus. J Mol Biol 337:1149–1160
Tomoike F, Nakagawa N, Kuramitsu S, Masui R (2011) A single amino acid limits the substrate specificity of Thermus thermophilus uridine-cytidine kinase to cytidine. Biochemistry 50:4597–4607
Welin M, Nordlund P (2010) Understanding specificity in metabolic pathways–structural biology of human nucleotide metabolism. Biochem Biophys Res Commun 396:157–163
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Driessen.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tomoike, F., Kuramitsu, S. & Masui, R. Unique substrate specificity of purine nucleoside phosphorylases from Thermus thermophilus . Extremophiles 17, 505–514 (2013). https://doi.org/10.1007/s00792-013-0535-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-013-0535-7