Abstract
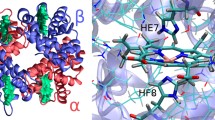
Cytoglobin (Cyg)—a new member of the vertebrate heme globin family—is expressed in many tissues of the human body but its physiological role is still unclear. It may deliver oxygen under hypoxia, serve as a scavenger of reactive species or be involved in collagen synthesis. This protein is usually six-coordinated and binds oxygen by a displacement of the distal HisE7 imidazole. In this paper, the results of 60 ns molecular dynamics (MD) simulations of dioxygen diffusion inside Cyg matrix are discussed. In addition to a classical MD trajectory, an approximate Locally Enhanced Sampling (LES) method has been employed. Classical diffusion paths were carefully analyzed, five cavities in dynamical structures were determined and at least four distinct ligand exit paths were identified. The most probable exit/entry path is connected with a large tunnel present in Cyg. Several residues that are perhaps critical for kinetics of small gaseous diffusion were discovered. A comparison of gaseous ligand transport in Cyg and in the most studied heme protein myoglobin is presented. Implications of efficient oxygen transport found in Cyg to its possible physiological role are discussed.

Pathways P3 (green), P3′ (red) and P3″ (blue). These pathways are located between G and H helices. Ligand moved from CavXe2Xe3 cavity to solvent using gateP3, gateP3′ or gateP3″. A detailed picture of the CavXe2Xe3 is available in Electronic Supplementary Materials (Fig. S13). CavXe2Xe3 constitute the main part of wide hydrophobic tunnel









Similar content being viewed by others

References
Kendrew JC (1963) Science 139:1259–1266
Perutz MF (1979) Annu Rev Biochem 48:327–386
Wittenberg JB (1970) Physiol Rev 50:559–636
Elber R, Karplus M (1987) Science 235:318–321
Wittenberg BA, Wittenberg JB (1989) Annu Rev Physiol 51:857–878
Perutz MF (1990) Annu Rev Physiol 52:1–25
Springer BA, Siligar SG, Olson JS, Philips GN (1994) Chem Rev 94:699–714
Tilton RF, Kuntz ID Jr, Petsko GA (1984) Biochem 23:2849–2857
Meller J, Elber R (1998) Biophys J 74:789–802
Burmester T, Weich B, Reinhardt S, Hankeln T (2000) Nature 407:520–523
Kawada N, Kristensen DB, Asahina K, Nakatani K, Minamiyama Y, Seki S, Yoshizato K (2001) J Biol Chem 276:25318–25323
Pesce A, Bolognesi M, Bocedi A, Ascenzi P, Dewilde S, Moens L, Hankeln T, Burmester T (2002) EMBO Rep 12:1146–1151
Burmester T, Ebner B, Weich B, Hankeln T (2002) Mol Biol Evol 19:416–421
Riggs A, Gorr T (2006) PNAS 103:2469–2470
Fordel E, Thijs L, Martinet W, Lenjou M, Laufs T, van Bockstaele D, Moens L, Dewilde S (2006) Neurosci Lett 10:146–151
Fago A, Hundahl C, Dewilde S, Gilany K, Moens L, Weber RE (2004) J Biol Chem 279:44417–44426
Schmid M, Gerlach F, Avivi A, Laufs T, Wystub S, Simpson JC, Nevoll E, Saaler-Reinhardt S, Reuss S, Hankeln T, Burmester T (2004) J Biol Chem 279:8063–8069
Hankeln T, Ebner B, Fuchs C, Gerlach F, Haberkamp M, Laufs TL, Roesner A, Schmidt M, Weich B, Wystub S, Saaler-Reinhardt S, Reuss S, Bolognesi M, De Sanctis D, Marden MC, Kiger L, Moens L, Dewilde S, Nevo E, Avivi A, Weber RE, Fago A, Burmester T (2005) J Inorg Biochem 99:110–119
Nakatani K, Okuyama H, Shimahara Y, Saeki S, Kim DH, Nakajima Y, Seki S, Kawada N, Yoshizato K (2004) Lab Invest 84:91
Dewilde S, Kieger L, Burmester T, Hankeln T, Baudin-Creuza V, Aerts T, Marden MC, Caubergs R, Moens L (2001) J Biol Chem 276:38949–38955
De Sanctis D, Dewilde S, Pesce A, Moens L, Ascenzi P, Hankeln T, Burmester T, Bolognesi M (2004) J Mol Biol 336:917–927
De Sanctis D, Dewilde S, Pesce A, Moens L, Ascenzi P, Hankeln T, Burmester T, Bolognesi M (2004) Biochem Biophys Res Commun 316:1217–1221
Nutt DR, Meuwly M (2004) Proc Natl Acad Sci USA 101:5998–6002
Bossa C, Anselmi M, Roccatano D, Amadei A, Vallone B, Brunori M, Di Nola A (2004) Biophys J 86:3855–3862
Bossa C, Amadei A, Daidone I, Anselmi M, Vallone B, Brunori M, Di Nola A (2005) Biophys J 89:465–474
Teeter MM (2004) Protein Sci 13:313–318
Cohen J, Arkhipov A, Braun R, Schulten K (2006) Biophys J 91:1844–1857
Elber R, Karplus MJ (1990) J Amer Chem Soc 112:9161–9175
Roitberg A, Elber R (1991) J Chem Phys 95:9277–9287
Verkhivker G, Elber R, Nowak W (1992) J Chem Phys 97:7838–7841
Nowak W, Czerminski R, Elber R (1991) J Amer Chem Soc 113:5627–5637
Sugimoto H, Makino M, Sawai H, Kawada N, Yoshizato K, Shiro Y (2004) J Mol Biol 339:873–885
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) Nucleic Acids Res 28:235–242
Kalr L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K (1999) J Comp Phys 151:283–312
Humphrey W, Dalke A, Schulten K (1996) J Molec Graphic 14:33–38
Guex N, Peitsch MC (1997) Electrophoresis 18:2714–2723
Orlowski S, Nowak W (2007) Theor Chem Acc 117:253–258
Karplus M, Straub J (1991) J Chem Phys 94:6737–6739
Scott EE, Gibson QH, Olson JS (2001) J Biol Chem 276:5177–5188
Schotte F, Lim M, Jackson TA, Smirnov AV, Soman J, Olson JS, Phillips GN, Wulff M, Anfinrud PA (2003) Science 300:1944–1947
Trent JT, Hargrove MS (2002) J Biol Chem 227:19538–19545
Brunori M, Cutruzzola F, Savino C, Travaglini-Allocatelli C, Vallone B, Gibson QH (1999) Biophys J 76:1259–1269
Tilton RF, Kuntz ID Jr, Petsko GA (1984) Biochemistry 23:2849–2857
Brunori M (2001) Trends Biochem Sci 26:209–210
Thompson JD, Higgins DG, Gibson TJ (1994) Nucleic Acids Res 22:4673–4680
Clamp M, Cuff J, Searle SM, Barton GJ (2004) Bioinformatics 20:426–427
Acknowledgements
This work was supported by grant “Krok w przyszlosc-stypendia dla doktorantow” (S.O.), President of Poland grant “Superpracownia 2002” and in part by MEiN grant 2P04A 07229 (W.N.). We thank CI TASK for computer time and Albert Rutkowski for his assistance in computations.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
RMSD analysis of all trajectories. The calculations were performed on Cα atoms using the initial structure as a reference. The blue plot represents the standard (LES1) trajectory. Black, red and green plots are from LES5, LES10 and LES15 trajectories, respectively (GIF 20 kb)
Fig. S2
Energy plots for the standard (LES1) trajectory, divided into total energy (black), kinetic (red) and potential (blue). The simulations are energetically stable (GIF 7 kb)
Fig. S3
Energy plots for the LES5 trajectory, divided into total energy (black), kinetic (red) and potential (blue). The simulations are energetically stable (GIF 7 kb)
Fig. S4
Energy plots for the LES10 trajectory, divided into total energy (black), kinetic (red) and potential (blue). The simulations are energetically stable (GIF 7 kb)
Fig. S5
Energy plots for the LES15 trajectory, divided into total energy (black), kinetic (red) and potential (blue). The simulations are energetically stable (GIF 7 kb)
Fig. S6
Temperature observed in the standard (LES1) trajectory. A value of the temperature achieved stabilization. The mean temperature is 299.72 ± 2.06 K (GIF 29 kb)
Fig. S7
Temperature observed in the LES5 trajectory. A value of the temperature achieved stabilization. The mean temperature is 299.74 ± 2.07 K (GIF 7 kb)
Fig. S8
Temperature observed in the LES10 trajectory. A value of the temperature achieved stabilization. The mean temperature is 299.74 ± 2.07 K (GIF 7 kb)
Fig. S9
Temperature observed in the LES15 trajectory. A value of the temperature achieved stabilization. The mean temperature is 299.74 ± 2.09 K (GIF 7 kb)
Fig. S10
Amino acids composition of the heme cavity (HemeCav). All phenylalanine residues are represented in red, leucines are displayed in green, Val85 in blue and His81 in yellow. The cytoglobin surface is represented in blue and the heme group with the proximal histidine is black (GIF 158 kb)
Fig. S11
All amino acids whose participate in building of the Xe1 cavity (CavXe1). All leucines are displayed in green and rest of the amino acids are properly labeled. The cytoglobin surface is represented in blue and the heme group with the proximal histidine is black (GIF 176 kb)
Fig. S12
All amino acids whose participate in building of the Xe4 cavity (CavXe4). The Cyg surface is represented in blue and the heme group with the proximal histidine is black (GIF 143 kb)
Fig. S13
All amino acids whose participate in building of the Xe2 Xe3 cavity (CavXe2Xe3). The color scheme is the same as on S10–S12 figures. Residue Ser128 is represented in pink, Val 93 is blue. The Cyg surface is represented in blue and the heme group with the proximal histidine is black (GIF 155 kb)
Fig. S14
All amino acids whose participate in building of the FE cavity (CavFE). The color scheme is the same as on previous figures. Residue Phe124 is represented in pink. The Cyg surface is represented in blue and the heme group with the proximal histidine is black (GIF 179 kb)
Rights and permissions
About this article
Cite this article
Orlowski, S., Nowak, W. Locally enhanced sampling molecular dynamics study of the dioxygen transport in human cytoglobin. J Mol Model 13, 715–723 (2007). https://doi.org/10.1007/s00894-007-0203-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-007-0203-x