Abstract
Isoproturon is the only herbicide that can control Phalaris minor, a competitive weed of wheat that developed resistance in 1992. Resistance against isoproturon was reported to be due to a mutation in the psbA gene that encodes the isoproturon-binding D1 protein. Previously in our laboratory, a triazole derivative of isoproturon (TDI) was synthesized and found to be active against both susceptible and resistant biotypes at 0.5 kg/ha but has shown poor specificity. In the present study, both susceptible D1(S), resistant D1(R) and D2 proteins of the PS-II reaction center of P. minor have been modeled and simulated, selecting the crystal structure of PS-II from Thermosynechococcus elongatus (2AXT.pdb) as template. Loop regions were refined, and the complete reaction center D1/D2 was simulated with GROMACS in lipid (1-palmitoyl-2-oleoylglycero-3-phosphoglycerol, POPG) environment along with ligands and cofactor. Both S and R models were energy minimized using steepest decent equilibrated with isotropic pressure coupling and temperature coupling using a Berendsen protocol, and subjected to 1,000 ps of MD simulation. As a result of MD simulation, the best model obtained in lipid environment had five chlorophylls, two plastoquinones, two phenophytins and a bicarbonate ion along with cofactor Fe and oxygen evolving center (OEC). The triazole derivative of isoproturon was used as lead molecule for docking. The best worked out conformation of TDI was chosen for receptor-based de novo ligand design. In silico designed molecules were screened and, as a result, only those molecules that show higher docking and binding energies in comparison to isoproturon and its triazole derivative were proposed for synthesis in order to get more potent, non-resistant and more selective TDI analogs.

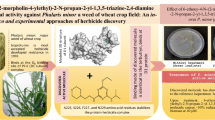
Pharmacophore of modeled and MD simulated (D1/D2) PS-II reaction center of Phalaris minor has been simulated for de novo synthesis of a TDI (triazole derivative of isoproturon)-based molecule. De novo synthesized molecules were redocked and reranked as more potent and selective herbicides for P. minor. The figure shows the modeled and MD simulated structure of the PS-II reaction center (D1(R)/D2) of P. minor (left), and the docked conformation of TDI in the binding cavity of D1(R) protein (right)







Similar content being viewed by others
Abbreviations
- MD:
-
Molecular dynamics simulation
- PS-II:
-
Photosystem-II
- D1/D2:
-
Photosystem second reaction center
- TDI:
-
Trizole derivative of isoproturon
- D1(R):
-
Resistant D1 protein
- D1(S):
-
Susceptible D1 protein
- POPG:
-
1-Palmitoyl-2-oleoylglycero-3-phosphoglycerol
- Tp:
-
Isotropic pressure coupling
- OEC:
-
Oxygen evolving center
- QB:
-
Plastoquinone B
- CDS:
-
Coding sequence
References
Bhan VM, Chaudhary DBS (1976) Germination, growth and reproducing behavior of Phalaris minor Retz as affected by the date of planting. Indian J Weed Sci 8:126–130
Malik RK, Singh S (1995) Little seed canary grass (Phalaris minor) resistance to isoproturon in India. Weed Technol 9:419–425
Walia US, Brar LS, Dhaliwal BK (1997) Resistant to isoproturon in Phalaris minor, Retz on Punjab. Plant Report 12:138–140
Yadav A, Malik RK, Balyan RS (1997) Studies on alternate herbicides to control isoproturon resistant littleseed canary grass. Pestology 21:26–28
Chhokar RS, Sharma RK (2008) Multiple herbicide resistant in little seed canary grass (Phalaris minor): a threat production in India Weed Bio Management 8:112–123
Singh DV, Gaur AK, Mishra DP (2004) Biochemical and molecular mechanism of resistance against isoproturon in Phalaris minor biotypes: variation in protein and RAPD profiles of isoproturon resistant and susceptible biotypes. Indian J Weed Sci 36:256–259
Tripathi MK, Yadav MK, Gaur AK, Mishra DP (2005) Resistant to isoproturon in Phalaris minor Retz on Punjab. Physiol Mol Biol Plants 11:161–163
Singh DV, Adeppa K, Misra K (2011) Mechanism of isoproturon resistance in Phalaris minor: in silico design, synthesis and testing of some novel herbicides for regaining sensitivity. J Mol Model. doi:10.1007/s00894-011-1169-2
Wu CH, Huang H, Nikolskaya A, Hu Z, Barker WC (2004) The iProClass integrated database for protein functional analysis. Comput Biol Chem 28:87–96
Loll B, Kern J, Zouni WA, Biesiadka J (2005) Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438:1040–1044
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Svensson B, Etchebest C, Tuffery P, van Kan P, Smith J, Styring S (1996) A model for the photosystem-II reaction center core including the structure of the primary donor P680. Biochemistry 35:14486–14502
Schlegel B, Sippl W, Holtje HD (2005) Molecular dynamics simulation of bovine rhodopsin: influence of protonation states and different membrane-mimicking environment. J Mol Model 12:49–64
Tieleman DP, Berendsen HJ (1996) Molecular dynamics simulation of fully hydrated DPPC with fully hydrated macroscopic boundary conditions and parameters. J Chem Phys 105:4871–4880
Astuti AD, Refianti R, Mutiara AB (2011) Molecular dynamics simulation on protein using GROMACS. Int J Comput Sci Inf Security 9:16–20
Ibragimova GT, Wade RC (1998) Stability of the beta-sheet of the WW domain: a molecular dynamics simulation study. Biophys J 74:2906–2911
Dundas J, Ouyang J, Tseng A, Binkowski Y, Turpaz LJ (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucl Acids Res 34:116–118
Morris GM, Goodsell DS, Huey R, Olson AJ (1996) Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2 4. J Comput Aided Mol Design 10:293–304
Wang R, Gao Y, Lai L (2000) LigBuilder: a multiple-purpose program for structure-based drug design. J Mol Model 6:498–516
Thomsen R, Christensen MH (2006) MolDock: a new technique for high-accuracy molecular docking. J Med Chem 49:3315–3321
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 1119 kb)
Rights and permissions
About this article
Cite this article
Singh, D.V., Agarwal, S., Kesharwani, R.K. et al. Molecular modeling and computational simulation of the photosystem-II reaction center to address isoproturon resistance in Phalaris minor . J Mol Model 18, 3903–3913 (2012). https://doi.org/10.1007/s00894-012-1386-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1386-3