Abstract
This study aimed to compare shallow and deep learning of classifying the patterns of interstitial lung diseases (ILDs). Using high-resolution computed tomography images, two experienced radiologists marked 1200 regions of interest (ROIs), in which 600 ROIs were each acquired using a GE or Siemens scanner and each group of 600 ROIs consisted of 100 ROIs for subregions that included normal and five regional pulmonary disease patterns (ground-glass opacity, consolidation, reticular opacity, emphysema, and honeycombing). We employed the convolution neural network (CNN) with six learnable layers that consisted of four convolution layers and two fully connected layers. The classification results were compared with the results classified by a shallow learning of a support vector machine (SVM). The CNN classifier showed significantly better performance for accuracy compared with that of the SVM classifier by 6–9%. As the convolution layer increases, the classification accuracy of the CNN showed better performance from 81.27 to 95.12%. Especially in the cases showing pathological ambiguity such as between normal and emphysema cases or between honeycombing and reticular opacity cases, the increment of the convolution layer greatly drops the misclassification rate between each case. Conclusively, the CNN classifier showed significantly greater accuracy than the SVM classifier, and the results implied structural characteristics that are inherent to the specific ILD patterns.



Similar content being viewed by others
References
Raghu G et al.: Incidence and prevalence of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 174(7):810–816, 2006
Scatarige JC et al.: Utility of high-resolution CT for management of diffuse lung disease: Results of a survey of US pulmonary physicians. Acad Radiol 10(2):167–175, 2003
Grenier P et al.: Chronic diffuse interstitial lung disease: Diagnostic value of chest radiography and high-resolution CT. Radiology 179(1):123–132, 1991
Kalender WA et al.: Measurement of pulmonary parenchymal attenuation: Use of spirometric gating with quantitative CT. Radiology 175(1):265–268, 1990
Chabat F, Yang G-Z, Hansell DM: Obstructive lung diseases: Texture classification for differentiation at CT 1. Radiology 228(3):871–877, 2003
Fujisaki T et al.: Effects of density changes in the chest on lung stereotactic radiotherapy. Radiat Med 22(4):233–238, 2003
Xu Y et al.: MDCT-based 3-D texture classification of emphysema and early smoking related lung pathologies. IEEE Trans Med Imaging 25(4):464–475, 2006
Delorme S et al.: Usual interstitial pneumonia: Quantitative assessment of high-resolution computed tomography findings by computer-assisted texture-based image analysis. Investig Radiol 32(9):566–574, 1997
Xu Y et al.: Computer-aided classification of interstitial lung diseases via MDCT: 3D adaptive multiple feature method (3D AMFM). Acad Radiol 13(8):969–978, 2006
Yuan R et al.: The effects of radiation dose and CT manufacturer on measurements of lung densitometry. Chest J 132(2):617–623, 2007
Lee Y et al.: Performance testing of several classifiers for differentiating obstructive lung diseases based on texture analysis at high-resolution computerized tomography (HRCT). Comput Methods Prog Biomed 93(2):206–215, 2009
Park YS et al.: Texture-based quantification of pulmonary emphysema on high-resolution computed tomography: Comparison with density-based quantification and correlation with pulmonary function test. Investig Radiol 43(6):395–402, 2008
Hoffman EA et al.: Characterization of the interstitial lung diseases via density-based and texture-based analysis of computed tomography images of lung structure and function 1. Acad Radiol 10(10):1104–1118, 2003
Uppaluri R et al.: Computer recognition of regional lung disease patterns. Am J Respir Crit Care Med 160(2):648–654, 1999
Wang J et al.: Computerized detection of diffuse lung disease in MDCT: The usefulness of statistical texture features. Phys Med Biol 54(22):6881, 2009
Yoon RG et al.: Quantitative assessment of change in regional disease patterns on serial HRCT of fibrotic interstitial pneumonia with texture-based automated quantification system. Eur Radiol 23(3):692–701, 2013
Park SO et al.: Comparison of usual interstitial pneumonia and nonspecific interstitial pneumonia: Quantification of disease severity and discrimination between two diseases on HRCT using a texture-based automated system. Korean J Radiol 12(3):297–307, 2011
Chang Y et al.: A support vector machine classifier reduces interscanner variation in the HRCT classification of regional disease pattern in diffuse lung disease: Comparison to a Bayesian classifier. Med Phys 40(5):051912, 2013
Krizhevsky A, Sutskever I, Hinton GE: Imagenet classification with deep convolutional neural networks. Adv Neural Inf Proces Syst 1097–1105, 2012
Mo D: A survey on deep learning: One small step toward AI. Albuquerque: Dept. Computer Science, Univ. of New Mexico, 2012
Goodfellow IJ et al.: Multi-digit number recognition from street view imagery using deep convolutional neural networks. arXiv preprint arXiv:1312.6082, 2013
Cruz-Roa AA et al.: A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013. Springer, 2013, pp 403–410
Bai W et al.: Multi-atlas segmentation with augmented features for cardiac MR images. Med Image Anal 19(1):98–109, 2015
de BrebissonA, Montana G: Deep Neural Networks for Anatomical Brain Segmentation. arXiv preprint arXiv:1502.02445, 2015
Li Q et al.: Medical image classification with convolutional neural network. Control Automation Robotics & Vision (ICARCV), 2014 13th International Conference on 844–848, 2014
Gao M et al.: Holistic Classification of CT Attenuation Patterns for Interstitial Lung Diseases via Deep Convolutional Neural Networks. crcv.ucf.edu
van Tulder G, de Bruijne M: Combining generative and discriminative representation learning for lung CT analysis with convolutional restricted boltzmann machines. IEEE Trans Med Imaging 35(5):1262–1272, 2016
Anthimopoulos M et al.: Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35(5):1207–1216, 2016
Shin H-C et al.: Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 35(5):1285–1298, 2016
Szegedy C et al.: Going deeper with convolutions. arXiv preprint arXiv:1409.4842, 2014
Acknowledgements
This study was supported by the Industrial Strategic Technology Development Program of the Ministry of Trade, Industry & Energy (10041618) in the Republic of Korea. This study is also the collaborated result supported by another national project of the ICT R&D program of MSIP/IITP (R6910-15-1023) in the Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Guk Bae Kim and Kyu-Hwan Jung are co-first authors.
Appendix
Appendix
Appendix 1
Appendix 2
Appendix 3
Appendix 4
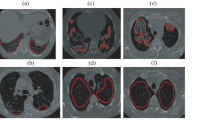
Comparison of whole lung quantification data using the golden standard by a radiologist (two cases). Each pixel was coded by the classification result, which is indicated by a semi-transparent color (normal, green; ground-glass opacity, yellow; reticular opacity, cyan; honeycombing, blue; emphysema, red; and consolidation, pink). a, d Original scanned images. b, e CNN classifier results. c, f Golden standard obtained by a radiologist. The colored areas beyond the lung were removed using a separately prepared lung mask
Rights and permissions
About this article
Cite this article
Kim, G.B., Jung, KH., Lee, Y. et al. Comparison of Shallow and Deep Learning Methods on Classifying the Regional Pattern of Diffuse Lung Disease. J Digit Imaging 31, 415–424 (2018). https://doi.org/10.1007/s10278-017-0028-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-017-0028-9