Abstract
Objectives
A potent HIV vaccine should overcome some limitations such as polymorphism of human HLA, the diversity of HIV-1 virus, and the lack of an effective delivery system. In this study, a DNA construct encoding Nef60–84, Nef126–144, Vpr34–47, Vpr60–75, Gp16030–53, Gp160308–323, and P248–151 epitopes was designed using bioinformatics tools. The pcDNA3.1-nef-vpr-gp160-p24 and pcDNA3.1-nef constructs were prepared in large scale as endotoxin-free form. Moreover, the recombinant Nef-Vpr-Gp160-p24 polypeptide and Nef protein were generated inE. coli. These constructs were delivered using cell penetrating peptides (CPPs) in vivo, and immune responses were assessed for different modalities in BALB/c mice.
Results
The recombinant DNA constructs were confirmed as the ~ 867 bp and ~ 648 bp bands related tonef-vpr-gp160-p24 andnef genes on agarose gel. Moreover, the purified Nef-Vpr-Gp160-p24 polypeptide and Nef protein showed the ~ 32 kDa and ~ 30 kDa bands on SDS-PAGE, respectively. The results of immune responses indicated that the heterologous prime/boost regimens using both Nef-Vpr-Gp160-P24 and Nef antigens induced significantly the secretion of IgG2a, IgG2b, IFN-γ and Granzyme B compared to other groups. The levels of Granzyme B in mice immunized with Nef antigen were higher than those immunized with Nef-Vpr-Gp160-P24 antigen. The CPPs showed the same potency with Montanide adjuvant for eliciting immune responses.
Conclusions
The heterologous prime/boost regimens for both antigens could significantly direct immune responses toward Th1 and CTL activity compared to other regimens. Comparing the efficiency of Nef-Vpr-Gp160-P24 and Nef constructs, the Nef-Vpr-Gp160-P24 constructs delivered by CPPs showed promising results as an HIV vaccine candidate.
Similar content being viewed by others
Introduction
Since the onset of the AIDS epidemic, 75 million people were infected with human immunodeficiency virus (HIV), and about 32 million died from HIV infection. At the end of 2018, about 37.9 million people were living with HIV worldwide (UNAIDS data 20192020). The HIV-1 virus is approximately 80–120 nm in diameter and was surrounded by a host plasma membrane (Karlsson et al.2008). The HIV-1 genome encodes nine proteins, which are divided into three categories: Gag, Pol and Env main structural proteins; Tat and Rev regulatory proteins; and Vif, Vpr, Vpu, and Nef accessory proteins (Goff2004).
Common vaccines, including live weakened or killed pathogens make up first-generation vaccines. Live weakened pathogens are usually much more effective and can induce humoral and cellular immune responses. However, concern about the safety of these vaccines prevents them from spreading and applying to diseases such as AIDS (Li and Petrovsky2015). In contrast, killed vaccines that are not dangerous cannot provide strong cellular immune responses and do not apply to all types of diseases. Therefore, to minimize the risks of first-generation vaccines, second-generation vaccines were developed as subunit vaccines. The new-generation vaccines are made from pathogenic parts such as protein antigens, synthetic peptides or polysaccharides. The limitations of this type of vaccines are their low ability to stimulate cellular immune responses and really low immunogenicity (Li and Petrovsky2015). Thus, the use of adjuvants and delivery systems, as well as heterologous prime/boost regimens was suggested to solve these problems. The studies indicated that repeated vaccine injections strengthen immune responses against the vaccine components. These strategies were known as prime/boost combination strategies. Initially, the immune system is stimulated by a vaccine candidate (prime) and re-stimulated with a similar (homologous) or different (heterologous) vaccine candidate with similar antigens as a reminder (boost) (Lu2009). For example, DNA prime/ protein boost regimens could be effective in stimulating immune responses. DNA vaccines have the potential to induce both cellular and humoral immune responses as the third generation of vaccines. However, their disadvantages can be reduced by heterologous prime/boost regimens and the use of delivery systems or adjuvants.
Montanide ISA-720 adjuvant is an example of a water/oil emulsion with an oil-to-water ratio of 70 to 30. The Montanide ISA-720 and its derivatives have been evaluated in various trials of malaria and AIDS vaccine. These adjuvants were very well tolerated when injected as a pharmaceutical formulation, and various clinical trials showed high tolerance of these compounds in combination with antigens. Various studies have demonstrated the ability of Montanide ISA-720 to enhance antibody titers as well as specific responses to cytotoxic T lymphocytes (CTL) (Aucouturier et al.2002). Moreover, delivery systems including cell penetrating peptides (CPPs) are a group of peptides that can cross different cell membranes with no lethal damage to the membrane (Jafari et al.2017). MPG and HR9 are examples of CPPs used to deliver DNA cargoes into the cells. MPG amphipathic peptide (27 amino acids) was composed of SV40 NLS and HIV glycoprotein 41 (gp41) separated by a linker (Wang et al.2014). Histidine-rich R9 (HR9, CH5-R9-H5C) contains polyhistidine and nona arginine (R9) sequences that are surrounded by two cysteine residues for increasing the delivery of DNA cargoes within the cells (Liu et al.2015; Lo and Wang2008; Huang et al.2015). On the other hand, a new cysteine-rich cationic CPP derived from crotamine known as CyLoP-1 has been recently developed to deliver peptide and protein cargoes (Ponnappan et al.2017; Jha et al.2011). LDP-NLS is another CPP containing Latarcin-derived peptide (LDP) and the nuclear localization sequence (NLS) of SV40 for delivery of protein and peptide cargoes into the cell (Ponnappan and Chugh2017; Dubovskii et al.2015; Ragin et al.2002).
Generally, finding an effective antigen and a suitable vaccine strategy is important to overcome HIV infection. In addition, safety of the designed constructs is critical for human use. Up to now, therapeutic vaccines achieved to clinical trials had some disadvantages (e.g., low immunogenicity, the lack of safety or high viral load). Thus, it is required to find novel vaccine strategies against HIV infection. The aim of this study was the design of a novel polyepitope construct and evaluation of its immunogenicity in mice. In our previous study, the pEGFP-nef-vpr-gp160-p24 construct and the Nef-Vpr-Gp160-P24 polypeptide complexed with CPPs were designed for in vitro studies (Davoodi et al.2019a,b). Herein, following in vitro studies, the pcDNA-nef-vpr-gp160-p24 and pcDNA-nef constructs, and the Nef-Vpr-Gp160-P24 polypeptide and Nef protein were prepared in large scale and endotoxin-free form. MPG & HR9 peptides and CyLoP-1 & LDP-NLS peptides were used to deliver DNA, and polypeptide/protein, respectively. Finally, humoral and cellular immune responses induced by different modalities using Nef-Vpr-Gp160-P24 and Nef antigens were evaluated and compared in BALB/c mice. Indeed, the efficiency of Nef-Vpr-Gp160-P24 regimens was compared with Nef regimens as an antigen candidate in HIV vaccine development. Briefly, the roles of CPPs, Montanide adjuvant and heterologous prime-boost strategy were evaluated to increase the immunogenicity of a polytope construct and a protein candidate in vivo. In addition, the potency of a polytope antigen against a protein antigen was determined to induce immune responses.
Materials and methods
Preparation of the polyepitope DNA construct
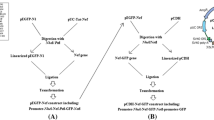
In our previous study (Davoodi et al.2019a,b), the Nef-Vpr-Gp160-P24 polyepitope peptide (/polypeptide) was designed by the conserved and immunogenic epitopes using bioinformatics analyses (Fig. 1). The molecular weight of the polypeptide was determined by the ExPASy website (https://web.expasy.org/compute_pi/). After reverse translation and codon optimization (https://www.bioinformatics.org/sms2/rev_trans.html), thenef-vpr-gp160-p24 DNA construct was ordered and synthesized in pUC57 cloning vector by Biomatik Corporation (Canada).
Cloning of the DNA construct in pcDNA3.1 (−)
The DNA construct (nef-vpr-gp160-p24) was digested from the pUC57 cloning vector usingBamHI andHindIII restriction enzymes (Thermo Fisher Scientific) and subcloned into theBamHI andHindIII cloning sites of pcDNA3.1(−) eukaryotic expression vector (Invitrogen) using T4 DNA ligase (Thermo Fisher Scientific). The pcDNA3.1 (−) vector harboring thenef-vpr-gp160-p24 gene construct was transformed into the competent DH5αE. coli using heat shock, and ampicillin-resistant colonies were selected. The pcDNA-nef-vpr-gp160-p24 was extracted using the Plasmid DNA Extraction Mini Kit (FAVORGEN, Taiwan) and purified by Xtra Maxi Plus Endotoxin-Free kit (MACHEREY-NAGEL, Germany). The concentration and purity of pcDNA-nef-vpr-gp160-p24 were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific). The presence of thenef-vpr-gp160-p24 gene was confirmed by digestion with the restriction enzymes and sequencing. The pcDNA3.1 (−) vector harboring the HIV-1nef gene was previously prepared by our group (Kadkhodayan et al.2017).
Preparation of the recombinant Nef-Vpr-Gp160-P24 polypeptide and Nef protein
The recombinant Nef-Vpr-Gp160-P24 polypeptide was expressed in theE. coli Rosetta strain transformed with pET-24a(+)-nef-vpr-gp160-p24 and purified by an affinity chromatography technique using Ni-NTA agarose column (Macherey-Nagel) under denaturing conditions as previously reported by our group (Davoodi et al.2019a,b). To produce Nef protein,Escherichia coli Rosetta strain was transformed with pET-23a (+) harboring thenef gene. Nef protein was expressed and purified under native conditions as previously reported by our group (Davoodi et al.2019a,b). In this study, the Nef-Vpr-Gp160-P24 polypeptide and Nef protein were produced in a large scale for mice immunization.
Preparation of the MPG/DNA and HR9/DNA nanoparticles
MPG (GALFLGFLGAAGSTMGAWSQPKKKRKV) (Morris1997) and HR9 (CHHHHHRRRRRRRRRHHHHHC) (Liu et al.2015) peptides were mixed with pcDNA-nef-vpr-gp160-p24 or pcDNA-nef at N/P (nitrogen to phosphate) ratios of 10 and 5, respectively for nanoparticles formation as previously reported (Davoodi et al.2019a,b). The size and charge of nanoparticles were assessed by scanning electron microscope (SEM) and Zetasizer at 25 °C, respectively.
Preparation of the CyLoP-1 or LDP-NLS/protein or polypeptide nanoparticles
CyLoP-1 (CRWRWKCCKK) (Jha et al.2011) and LDP-NLS (CHHHHHRRRRRRRRRHHHHHC) (Ponnappan and Chugh2017) peptides were mixed with Nef-Vpr-Gp160-P24 polypeptide and Nef protein at a molar ratio of 10:1 for nanoparticles formation as previously reported (Davoodi et al.2019a,b).
Mice immunization
Six to eight-week-old female BALB/c mice were provided from breeding stock maintained at Pasteur Institute of Iran. All mice were maintained under pathogen-free conditions. The whole process was performed based on approved protocols and care of laboratory animals at Pasteur Institute of Iran. Fifteen groups of mice (n = 4 mice per group) were considered and immunized subcutaneously at the footpad three times at 2-week intervals for both polypeptide and protein. The injectednef-vpr-gp160-p24 andnef DNA constructs (5 µg) and also the Nef-Vpr-Gp160-P24 polypeptide and Nef protein (5 µg) were diluted in endotoxin-free PBS 1X. The Nef-Vpr-Gp160-P24 polypeptide and Nef protein were emulsified with Montanide ISA-720 at the ratio of 70:30 (v/v, oil: aqueous phase). Three different regimens were used for mice immunization including DNA prime/DNA boost (homologous), protein or polypeptide prime/protein or polypeptide boost (homologous), and DNA prime/protein or polypeptide boost (heterologous) regimens (Tables 1 and2).
Antibody assay
Four weeks after the third injection, the sera of mice were collected and pooled for each group and stored at − 20 °C. The levels of total IgG, IgG1, IgG2a and IgG2b (1:10,000 v/v, Sigma) were assessed by indirect ELISA against the Nef-Vpr-Gp160-P24 polypeptide or Nef protein as an antigen (5 µg/ml). TMB (Tetra Methyl Benzidine, Sigma) was used as a substrate and the absorbance was assessed at 450 nm (Bolhassani et al.2008).
Cytokine assay
Four weeks after the third injection, three mice from each group were sacrificed and the spleens were removed. The red blood cell-depleted splenocytes pooled for each group (2 × 106 cells/ml) were seeded in 48-well plates (Nunc, Germany) in the presence of 5 µg/ml of Nef-Vpr-Gp160-P24 polypeptide or Nef protein, RPMI 5% (negative control), and 5 µg/ml of concanavalin A (ConA, positive control). The rest of the splenocytes were simultaneously used to assess the Granzyme B release. The levels of IFN-γ, IL-5 and IL-10 in supernatants were measured using a sandwich-based ELISA system (Mabtech, Sweden) according to the manufacturer’s instructions. The detection limit was 4 pg/ml for IFN-γ, 1 pg/ml for IL-5 and 4 pg/ml for IL-10.
Granzyme B assay
SP2/0 target cells (T) were cultured in triplicate into U-bottomed, 96-well plates (2 × 104 cells per well) incubated with Nef-Vpr-Gp160-P24 or Nef antigen (∼ 5 µg/ml) for a day. Then, the splenocyte effector cells (E) were added to the target cells at E:T ratio of 100:1 (maximal release of Granzyme B) and incubated for 6 h. Finally, the supernatants were collected, and the concentration of Granzyme B was measured in each sample by ELISA kit (eBioscience, USA) according to the manufacturer’s instruction.
Statistical analysis
The differences between the control and test groups were assessed using one-way ANOVA (Graph-pad Prism, GraphPad Software). Ap value < 0.05 was considered statistically significant. Results were expressed as mean ± standard deviation. Similar results were obtained in two independent experiments.
Results
Construction of the recombinant pcDNA- nef-vpr-gp160-p24 vector
At first, based on immunoinformatics analysis, the polyepitope peptide (/polypeptide) construct was designed using the selected Nef60–84, Nef126–144, Vpr34–47, Vpr60–75, Gp16030–53, Gp160308–323 and P248–151 epitopes (Fig. 1). Then, thenef-vpr-gp160-p24 DNA construct was subcloned into the pcDNA3.1 (−) vector. The presence of thenef-vpr-gp160-p24 gene in pcDNA3.1 (−) was confirmed by enzymatic digestion with BamHI and HindIII, and also sequencing. Thenef-vpr-gp160-p24 gene was observed as a clear band of ~ 867 bp in 1% agarose gel (Fig. 2). The concentration of the pcDNA-nef-vpr-gp160-p24 construct purified using an Endotoxin-free kit was 2250 ng/µl.
Subcloning of the DNA construct in pcDNA3.1(−):a Lane 1: double digested pUC57 (~ 2710 bp) andnef-vpr-gp160-p24 construct (~ 867 bp) using BamHI and HindIII restriction enzymes, Lane 2: pUC57-nef-vpr-gp160-p2 plasmid (~ 3541 bp);b Lane 1: Double digested pcDNA3.1(−) (~ 5427 bp) andnef-vpr-gp160-p24 construct (~ 867 bp) using BamHI and HindIII restriction enzymes, Lane 2: The purified pcDNA-nef-vpr-gp160-p24 (~ 6294 bp); MW: Molecular size marker (DNA ladder, 1 kb, SMOBIO)
Generation of the recombinant polypeptide/protein in large scale
The Nef-Vpr-Gp160-P24 polypeptide was expressed in Rosetta strain at 37 °C and 4 h after IPTG induction, and purified under denaturing conditions as a clear band of ~ 32 kDa confirmed in SDS-PAGE. Also, the Nef protein was expressed in Rosetta at 37 °C and 16 h after induction and purified under native conditions as a clear band of ~ 30 kDa confirmed in SDS-PAGE.
Preparation of the CPPs/DNA, and CPPs/protein or polypeptide nanoparticles
MPG/pcDNA-nef-vpr-gp160-p24, MPG/pcDNA-nef, HR9/pcDNA-nef-vpr-gp160-p24, and HR9/pcDNA-nef gel retardation assay showed that DNA did not migrate in agarose gel at N/P ratios of 10, 10, 5 and 5, respectively indicating the formation of complexes. SEM results indicated that spherical MPG/pcDNA-nef-vpr-gp160-p24 and MPG/pcDNA-nef complexes and nonspherical HR9/pcDNA-nef-vpr-gp160-p24 and HR9/pcDNA-nef complexes had an average diameter of about 100–200 nm. Zetasizer results showed that all mentioned complexes had positive charges between + 22 and + 26 mV. On the other hand, SEM data indicated that spherical CyLoP-1/Nef-Vpr-Gp160-P24 and CyLoP-1/Nef complexes and nonspherical LDP-NLS/Nef-Vpr-Gp160-P24 and LDP-NLS/Nef complexes had an average diameter of about 100–200 nm at molar ratio of 10: 1 (CPP: protein or peptide). Zetasizer data demonstrated that all mentioned complexes had positive charges between + 4 and + 10 mV.
Antibody assay
The results of total IgG and its isotypes indicated that the difference between the test groups (G1–G12) and control groups (G13–G15) was statistically significant (p < 0.001) for both Nef-Vpr-Gp160-P24 and Nef antigens (Figs. 3,4). The levels of total IgG in mice immunized with homologous protein or polypeptide regimens (G4–G6) were significantly higher (p < 0.001) than heterologous DNA prime/ protein or polypeptide boost regimens (Figs. 3a,4a). The levels of IgG1 in mice immunized with protein or polypeptide emulsified by Montanide (G4) were significantly higher (p < 0.001) than other regimens (Figs. 3b,4b). The levels of IgG2a in mice immunized with heterologous regimens (G7–G12) were significantly higher (p < 0.001) than homologous regimens for both antigens (Figs. 3c,4c). The levels of IgG2b in mice immunized with homologous polypeptide regimens and heterologous DNA/ polypeptide regimens (G4–G12) were significantly higher (p < 0.001) than homologous DNA regimens (G1–G3) (Fig. 3d). The levels of IgG2b in mice immunized with heterologous DNA/ protein regimens (G7–G12) were significantly higher (p < 0.001) than homologous protein regimen (Fig. 4d). Overall, the levels of total IgG and IgG2b in mice immunized with Nef-Vpr-Gp160-P24 polypeptide and Nef protein showed no significant differences (p > 0.05). The levels of IgG1 and IgG2a in mice immunized with Nef antigen were higher than those immunized with Nef-Vpr-Gp160-P24 antigen. The ratios of IgG2a/IgG1 and IgG2b/IgG1 were approximately 3 and 2, respectively for both antigens suggesting direction toward Th1 response.
Antibody responses against Nef-Vpr-Gp160-P24 antigen in different regimens:a Total IgG,b IgG1,c IgG2a andd IgG2b. All analyses were performed in duplicate for each sample. The results from the 1:100 sera dilutions were shown as mean absorbance at 450 nm ± SD. Significant differences were shown by *p < 0.05, **p < 0.01, ***p < 0.001, and non-significant difference was shown by ns (p > 0.05)
Antibody responses against Nef antigen in different regimens:a Total IgG,b IgG1,c IgG2a andd IgG2b. All analyses were performed in duplicate for each sample. The results from the 1:100 sera dilutions are shown as mean absorbance at 450 nm ± SD. Significant differences were shown by *p < 0.05, **p < 0.01, ***p < 0.001, and non-significant difference was shown by ns (p > 0.05)
Cytokine assay
The results of cytokine assay indicated that the difference between the test groups (G1–G12) and control groups (G13–G15) was statistically significant for both Nef-Vpr-Gp160-P24 and Nef antigens (p < 0.001) (Figs. 5,6). The levels of IFN-γ in mice immunized with heterologous regimens (G7–G12) were significantly higher (p < 0.001) than homologous regimens for both antigens (Nef-Vpr-Gp160-P24 and Nef) (Figs. 5a,6a). The levels of IL-5 in mice immunized with homologous protein or polypeptide regimens (G4–G6) and heterologous DNA/protein or polypeptide regimens (G7–G12) were significantly higher (p < 0.05) than the homologous DNA regimens (Figs. 5b, 6b). The levels of IL-10 in mice immunized with homologous polypeptide regimens mixed with Montanide (G4) and CyLoP-1 (G5) were significantly higher (p < 0.001) than other regimens (Fig. 5c). The levels of IL-10 in mice immunized with homologous Nef protein regimen mixed with Montanide (G4) were significantly higher (p < 0.05) than other regimens (Fig. 6c). Overall, the levels of IFN-γ in mice immunized with Nef-Vpr-Gp160-P24 and Nef showed no significant differences (p > 0.05). The levels of IL-5 and IL-10 in mice immunized with Nef-Vpr-Gp160-P24 were higher than those immunized with Nef (p < 0.05). However, the ratios of IFN-γ/IL-5 and IFN-γ/IL-10 were approximately 14 and 5, respectively for both antigens suggesting direction toward Th1 response.
Cytokine secretion in mice immunized with Nef-Vpr-Gp160-P24 in different regimens:a IFN-γ,b IL-5 andc IL-10. The level of cytokines was determined in the supernatant with ELISA as mean absorbance at 450 nm ± SD for each set of samples. All analyses were performed in duplicate for each sample. Significant differences were shown by *p < 0.05, **p < 0.01, ***p < 0.001, and non-significant difference was shown by ns (p > 0.05)
Cytokine secretion in mice immunized with Nef in different regimens:a IFN-γ,b IL-5 andc IL-10. The level of cytokines was determined in the supernatant with ELISA as mean absorbance at 450 nm ± SD for each set of samples. All analyses were performed in duplicate for each sample. Significant differences were shown by *p < 0.05, **p < 0.01, ***p < 0.001, and non-significant difference was shown by ns (p > 0.05)
Granzyme B assay
The results of Granzyme B assay indicated that the difference between test (G1–G12) and control (G13–G15) groups was significant (p < 0.001) for both Nef-Vpr-Gp160-P24 and Nef antigens (Fig. 7). The levels of Granzyme B in mice immunized with heterologous regimens (G7–G12) were significantly higher (p < 0.01) than homologous regimens for both antigens (Nef-Vpr-Gp160-P24 and Nef) (Fig. 7a, b). Overall, the levels of Granzyme B in mice immunized with Nef were higher than those immunized with Nef-Vpr-Gp160-P24 as a possible CTL activity (p < 0.05).
Granzyme B secretion in mice immunized with Nef-Vpr-Gp160-P24 (a) and Nef (b) in different regimens. The level of Granzyme B was determined with ELISA as mean absorbance at 450 nm ± SD for each set of samples. All analyses were performed in duplicate for each sample. Significant differences were shown by *p < 0.05, **p < 0.01, ***p < 0.001, and non-significant difference was shown by ns (p > 0.05)
Discussion
In this study, we evaluated the efficiency of a new polyepitope construct as an antigen candidate along with delivery systems and adjuvant for inducing effective immune responses. The immunogenicity of the designed construct was compared with Nef protein as an antigen candidate in therapeutic vaccine. Some studies have focused on Nef, a highly conserved and immunogenic regulatory protein in vaccine design (Mann and Ndungu2015; Abraham and Fackler2012; Jafarzade et al.2016; Kadkhodayan et al.2017). In addition, many of the completed HIV-1 vaccine experiments have used recombinant viral vectors or DNA vaccines in combination with other traditional vector-based vaccines. Immune responses primed by injection of an antigen-encoding DNA could be enhanced using recombinant proteins or viruses through “prime/boost” strategies. In animal models, these approaches have increased the number of neutralizing antibodies and DNA-based CTL responses (Kutzler and Weiner2008). Several studies used a set of proteins as multivalent immunogens in vaccine design. One of these strategies was the bioinformatics design of artificial proteins that could cover different epitopes. The challenge for these strategies was to design vaccines that could provide protective responses against the highly diverse range of HIV-1 viruses (Korber et al.2017). So far, several peptide vaccines including HIV-v (Vpr, Vif, Rev, Nef), C4-V3 (Gp120), VAC3S (Gp41), VACC-4X (p24Gag), F4/AS01B (p24, RT, Nef, p17) and Afo-18 have been developed by various scientists, but the capacity of these vaccines has also been limited to increase HIV-specific immune responses (Leal et al.2017).
In this study, a DNA construct was designed based on the conserved epitopes of HIV-1 subtypes which interacts with common HLAs in the world. These immunodominant epitopes include Nef60–84, Nef126–144, Vpr34–47, Vpr60–75, Gp16030–53, Gp160308–323, and P248–151 and the interaction between these epitopes and common HLAs was investigated using CABS-dock peptide-protein docking website. Our main approach was the stimulation of cellular immunity and cytotoxic T lymphocytes. Therefore, the epitopes were selected to simultaneously stimulate CD4+ (T helper) and CD8+ (cytotoxic T) T cells based on online softwares. Herein, it seems that the simultaneous use of the mentioned epitopes increases vaccine success. Moreover, due to the increase in the length of the polyepitope peptide, two main drawbacks of peptide vaccines including low immunogenicity and MHC limitation were eliminated because they acquire the characteristics of protein vaccines without areas that are not immunogenic or do not have effective immunogenicity. Elimination of non-immunogenic components also reduces the side effects of the vaccine (Purcell et al.2007). On the other hand, many studies have focused on increasing immunogenicity by using immune stimulants or adjuvants. Adjutants act by activating the innate immune system and providing key signals that regulate the acquired immune response. As a result, they stimulate antigen-specific T helper cells (Th1, Th2, and Th17) and ultimately lead to immunity (Bonam et al.2017; Draper et al.2015). In this study, the Montanide ISA-720 adjuvant was used. This adjuvant has an FDA license for use in human vaccines (Tsang et al.2017).
The main drawback of DNA vaccines is their low immunogenicity in clinical trials. DNA vaccines are easily destroyed by DNase and lysozyme, and the injected naked plasmid DNA is poorly distributed and inefficiently expressed. Therefore, DNA vaccines must be injected by a delivery system to increase immune responses (Tian et al.2014). Cell-penetrating peptides were widely used to deliver proteins, plasmid DNAs, RNAs, oligonucleotides, liposomes, and anticancer drugs into the cells (Borrelli et al.2018). CPPs are generally bound to biomolecules by covalent binding or forming non-covalent complexes (Kawamoto et al.2011; Simeoni2003). The important benefits of forming non-covalent complexes are the ease of complex preparation and conservation of cargo characteristics (Huang et al.2015). In this study, four cell penetrating peptides were used to effectively deliver the DNA, polypeptide or protein constructs. We used three prime/boost vaccine strategies including DNA prime/DNA boost (homologous), protein prime/protein or polypeptide boost (homologous), and DNA prime/protein or polypeptide boost (heterologous) strategies. The best efficiency of immune responses was observed in the heterologous (G7–G12) and homologous protein/polypeptide groups (G4–G6) compared to homologous DNA groups (G1–G3). An increase in the ratios of IgG2a and IgG2b to IgG1 in heterologous groups indicated the tendency of the immune responses to Th1 (cellular immunity). Moreover, an increase in the ratios of IFN-γ to IL-5 and IL-10 cytokines in these groups showed the direction of the immune responses to Th1 (cellular immunity). Increased production of Granzyme B in these groups was also an indicator of CTL activity. Indeed, in the heterologous prime/boost groups, the highest immune response was related to IgG2a, IgG2b, and IFN-γ (tendency to Th1) for both antigens. Generally, the use of three strategies including heterologous prime/boost, MPG/HR9 for DNA delivery and CyLoP-1/LDP-NLS for protein or polypeptide delivery could significantly activate both T- and B-cells against both antigens.
In various studies, the efficiency of prime/boost strategy has been proved compared to other strategies. One research showed that the heterologous DNA prime/ peptide boost strategy using HIV-1-MPER-V3 antigen was more effective than homologous strategies in eliciting humoral and cellular immune responses. Besides, the use of MPG as a DNA delivery system increased the immunological efficiency of DNA construct (Kardani et al.2016). In another study, Kadkhodayan and colleagues indicated that the heterologous HIV-1 Tat (CPP)-Nef DNA prime/ HIV-1 Tat (CPP)-Nef protein boost strategy increased IgG2a, IFN-γ, and Granzyme B secretion in BALB/c mice. They used Cady-2 cell penetrating peptide as a non-covalent protein delivery system as well as Tat CPP as a covalent delivery system (Kadkhodayan et al.2017). In the current study, the results of immune responses showed that the heterologous prime/boost regimens using both Nef-Vpr-Gp160-P24 and Nef antigens induced significantly the secretion of IgG2a, IgG2b, IFN-γ and Granzyme B in comparison with other groups. The levels of Granzyme B in mice immunized with Nef antigen were higher than those immunized with Nef-Vpr-Gp160-P24 antigen. The CPPs showed the same potency with Montanide adjuvant for eliciting immune responses.
In conclusion, a polyepitope DNA construct was designed using bioinformatics tools which had the conserved immunogenic epitopes of HIV-1 subtypes and could enhance humoral and cellular immune responses. In addition, we attempted to express a recombinant polyepitope peptide as an antigen candidate. Indeed, we synthesized the polyepitope DNA construct and then express it in bacterial system. As known, a recombinant polypeptide is more efficient than a synthetic peptide for inducing effective immune responses. We used a proteasomal linker between each epitope for cutting and presentation by MHC-I class. On the other hand, cell-penetrating peptides were used to deliver DNA and polypeptide constructs into the cells. The results of humoral and cellular immune responses showed that heterologous prime/boost groups and then homologous protein groups using Nef-Vpr-Gp160-P24 polypeptide and Nef protein had more significant immune responses than homologous DNA groups (p < 0.05). The secretion of IgG2a, IgG2b, IFN-γ and Granzyme B in heterologous prime/boost groups along with the use of adjuvant and delivery systems was higher than other groups indicating a greater tendency toward Th1 and CTL responses (p < 0.05). These results showed that Nef-Vpr-Gp160-P24 polypeptide has similar efficiency with Nef protein as an antigen candidate in eliciting immune responses for HIV vaccine development. However, this polyepitope construct is more effective than Nef protein due to the conservancy and low side effects for using in clinical trials.
References
Abraham L, Fackler O (2012) HIV-1 Nef: a multifaceted modulator of T cell receptor signaling. Cell Commun Signal 10(1):39.https://doi.org/10.1186/1478-811x-10-39
Aucouturier J, Dupuis L, Deville S, Ascarateil S, Ganne V (2002) Montanide ISA 720 and 51: a new generation of water in oil emulsions as adjuvants for human vaccines. Expert Rev Vaccines 1(1):111–118.https://doi.org/10.1586/14760584.1.1.111
Bolhassani A, Zahedifard F, Taghikhani M, Rafati S (2008) Enhanced immunogenicity of HPV16 E7 accompanied by Gp96 as an adjuvant in two vaccination strategies. Vaccine 26(26):3362–3370.https://doi.org/10.1016/j.vaccine.2008.03.082
Bonam S, Partidos C, Halmuthur S, Muller S (2017) An overview of novel adjuvants designed for improving vaccine efficacy. Trends Pharmacol Sci 38(9):771–793.https://doi.org/10.1016/j.tips.2017.06.002
Borrelli A, Tornesello A, Tornesello M, Buonaguro F (2018) Cell penetrating peptides as molecular carriers for anti-cancer agents. Molecules 23(2):295.https://doi.org/10.3390/molecules23020295
Davoodi S, Bolhassani A, Sadat S, Irani S (2019a) Design and in vitro delivery of HIV-1 multi-epitope DNA and peptide constructs using novel cell-penetrating peptides. Biotechnol Lett 41(11):1283–1298.https://doi.org/10.1007/s10529-019-02734-x
Davoodi S, Bolhassani A, Sadat S, Irani S (2019b) Enhancing HIV-1 Nef penetration into mammalian cells as an antigen candidate. J Med Microbiol Infect Dis 7(1):37–43.https://doi.org/10.29252/jommid.7.1.2.37
Draper S, Angov E, Horii T et al (2015) Recent advances in recombinant protein-based malaria vaccines. Vaccine 33(52):7433–7443.https://doi.org/10.1016/j.vaccine.2015.09.093
Dubovskii P, Vassilevski A, Kozlov S, Feofanov A, Grishin E, Efremov R (2015) Latarcins: versatile spider venom peptides. Cell Mol Life Sci 72(23):4501–4522.https://doi.org/10.1007/s00018-015-2016-x
Goff S (2004) Retrovirus restriction factors. Mol Cell 16(6):849–859.https://doi.org/10.1016/j.molcel.2004.12.001
Huang Y, Lee H, Tolliver L, Aronstam R (2015) Delivery of nucleic acids and nanomaterials by cell-penetrating peptides: opportunities and challenges. Biomed Res Int.https://doi.org/10.1155/2015/834079
Jafari S, Maleki Dizaj S, Adibkia K (2017) Cell-penetrating peptides and their analogues as novel nanocarriers for drug delivery. Bioimpacts 5(2):103–111.https://doi.org/10.15171/bi.2015.10
Jafarzade B, Bolhassani A, Sadat S, Yaghobi R (2016) Delivery of HIV-1 Nef protein in mammalian cells using cell penetrating peptides as a candidate therapeutic vaccine. Int J Pept Res Ther 23(1):145–153.https://doi.org/10.1007/s10989-016-9547-3
Jha D, Mishra R, Gottschalk S et al (2011) CyLoP-1: A novel cysteine-rich cell-penetrating peptide for cytosolic delivery of cargoes. Bioconjug Chem 22(3):319–328.https://doi.org/10.1021/bc100045s
Kadkhodayan S, Jafarzade B, Sadat S, Motevalli F, Agi E, Bolhassani A (2017) Combination of cell penetrating peptides and heterologous DNA prime/protein boost strategy enhances immune responses against HIV-1 Nef antigen in BALB/c mouse model. Immunol Lett 188:38–45.https://doi.org/10.1016/j.imlet.2017.06.003
Kardani K, Bolhassani A, Shahbazi S (2016) Prime-boost vaccine strategy against viral infections: mechanisms and benefits. Vaccine 34(4):413–423.https://doi.org/10.1016/j.vaccine.2015.11.062
Karlsson HG, Fouchier R et al (2008) The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Microbiol 6(2):143–155.https://doi.org/10.1038/nrmicro1819
Kawamoto S, Takasu M, Miyakawa T et al (2011) Inverted micelle formation of cell-penetrating peptide studied by coarse-grained simulation: importance of attractive force between cell-penetrating peptides and lipid head group. J Chem Phys 134(9):095103.https://doi.org/10.1063/1.3555531
Korber B, Hraber P, Wagh K, Hahn B (2017) Polyvalent vaccine approaches to combat HIV-1 diversity. Immunol Rev 275(1):230–244.https://doi.org/10.1111/imr.12516
Kutzler M, Weiner D (2008) DNA vaccines: ready for prime time? Nat Rev Genet 9(10):776–788.https://doi.org/10.1038/nrg2432
Leal L, Lucero C, Gatell J, Gallart T, Plana M, García F (2017) New challenges in therapeutic vaccines against HIV infection. Expert Rev Vaccines 16(6):587–600.https://doi.org/10.1080/14760584.2017.1322513
Li L, Petrovsky N (2015) Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev Vaccines 15(3):313–329.https://doi.org/10.1586/14760584.2016.1124762
Liu B, Chen H, Chan M, Huang Y, Aronstam R, Lee H (2015) Three arginine-rich cell-penetrating peptides facilitate cellular internalization of red-emitting quantum dots. J Nanosci Nanotechnol 15(3):2067–2078.https://doi.org/10.1166/jnn.2015.9148
Lo S, Wang S (2008) An endosomolytic Tat peptide produced by incorporation of histidine and cysteine residues as a nonviral vector for DNA transfection. Biomaterials 29(15):2408–2414.https://doi.org/10.1016/j.biomaterials.2008.01.031
Lu S (2009) Heterologous prime-boost vaccination. Curr Opin Immunol 21(3):346–351.https://doi.org/10.1016/j.coi.2009.05.016
Mann J, Ndungu T (2015) HIV-1 vaccine immunogen design strategies. Virol J 12(1):3.https://doi.org/10.1186/s12985-014-0221-0
Morris M (1997) A new peptide vector for efficient delivery of oligonucleotides into mammalian cells. Nucleic Acids Res 25(14):2730–2736.https://doi.org/10.1093/nar/25.14.2730
Ponnappan N, Chugh A (2017) Cell-penetrating and cargo-delivery ability of a spider toxin-derived peptide in mammalian cells. Eur J Pharm Biopharm 114:145–153.https://doi.org/10.1016/j.ejpb.2017.01.012
Ponnappan N, Budagavi D, Chugh A (2017) CyLoP-1: membrane-active peptide with cell-penetrating and antimicrobial properties. Biochimica et Biophysica Acta (BBA) 1859(2):167–176.https://doi.org/10.1016/j.bbamem.2016.11.002
Purcell A, McCluskey J, Rossjohn J (2007) More than one reason to rethink the use of peptides in vaccine design. Nat Rev Drug Discov 6(5):404–414.https://doi.org/10.1038/nrd2224
Ragin A, Morgan R, Chmielewski J (2002) Cellular import mediated by nuclear localization signal peptide sequences. Chem Biol 9(8):943–948.https://doi.org/10.1016/s1074-5521(02)00189-8
Simeoni F (2003) Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res 31(11):2717–2724.https://doi.org/10.1093/nar/gkg385
Tian Y, Wang H, Liu Y et al (2014) A peptide-based nanofibrous hydrogel as a promising DNA nanovector for optimizing the efficacy of HIV vaccine. Nano Lett 14(3):1439–1445.https://doi.org/10.1021/nl404560v
Tsang K, Fantini M, Fernando R et al (2017) Identification and characterization of enhancer agonist human cytotoxic T-cell epitopes of the human papillomavirus type 16 (HPV16) E6/E7. Vaccine 35(19):2605–2611.https://doi.org/10.1016/j.vaccine.2017.03.025
UNAIDS data 2019 (2020)http://rstesa.unaids.org/publications/global-publications/item/208-unaids-data-2019.. Accessed 2 Jan 2020
Wang F, Wang Y, Zhang X, Zhang W, Guo S, Jin F (2014) Recent progress of cell-penetrating peptides as new carriers for intracellular cargo delivery. J Controlled Release 174:126–136.https://doi.org/10.1016/j.jconrel.2013.11.020
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Six to eight-week-old female BALB/c mice were provided from breeding stock maintained at Pasteur Institute of Iran. All mice were maintained under pathogen-free conditions. The whole mice experiments were done based on approved protocols for care of laboratory animals at Pasteur Institute of Iran (a local approval Committee; ethical code No: IR.PII.REC.1397.024).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Davoodi, S., Bolhassani, A. & Namazi, F. In vivo delivery of a multiepitope peptide and Nef protein using novel cell-penetrating peptides for development of HIV-1 vaccine candidate. Biotechnol Lett 43, 547–559 (2021). https://doi.org/10.1007/s10529-020-03060-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-020-03060-3