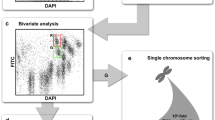
Abstract
Flow-sorted chromosomes have been used to simplify analyses of complex plant genomes. In bread wheat, majority of studies involve cultivar Chinese Spring, a genotype chosen for sequencing. Telosomic lines developed from this cultivar enable isolation by flow sorting chromosome arms, which represent less than 3.4 % of the genome. However, access to other wheat cultivars is needed to allow mapping and cloning useful genes. In these cultivars, cytogenetic stocks are not readily available and only one chromosome (3B) can be sorted. Remaining chromosomes form composite peaks on flow karyotypes and cannot be sorted. In order to overcome this difficulty, we tested a pragmatic approach in which composite chromosome peaks are dissected to smaller sections. The analysis of chromosome composition in sorted fractions confirmed feasibility of obtaining sub-genomic fractions comprising only a few chromosomes. Usually one of the chromosomes was more abundant and the frequencies of dominant chromosomes in sorted fractions ranged from 16 % (chromosome 7B) to 80 % (chromosome 2B). The enrichment factor, calculated as the relative proportion of chromosomal DNA in the wheat genome to the proportion of chromosomal DNA in a sorted fraction, ranged from 3.2-fold (7B) to 16.4-fold (5D). At least a 5-fold enrichment can be obtained for 17 out of 21 wheat chromosomes. Moreover, we show that 15 out of the 21 chromosomes can be sorted without being contaminated by their homoeologs. These observations provide opportunities for constructing sub-genomic large-insert DNA libraries, optical mapping, and targeted sequencing selected genome regions in various cultivars of wheat. The availability of fractions enriched for chromosomes of interest and free of contaminating homoeologs will increase the efficiency of research projects and reduce their costs as compared to whole genome approaches. The same methodology should be feasible in other plants where single chromosome types cannot be sorted.
Similar content being viewed by others
Abbreviations
- DAPI:
-
4′,6-diamidino-2-phenylindole
- FISH:
-
fluorescence in situ hybridization
- FISHIS:
-
FISH in suspension
- PRINSES:
-
primed in situ labelling en suspension
- RIL:
-
recombinant inbred line
- STS:
-
sequence-tagged site
References
Bedbrook, J.R., Jones, J., O’Dell, M., Thompson, R.D., Flavell, R.B.: A molecular description of telomeric heterochromatin in Secale species. — Cell 19: 545–560, 1980.
Bennetzen, J.L.: Transposable element contributions to plant gene and genome evolution. — Plant mol. Biol. 42: 251–269, 2000.
Doležel, J., Kubaláková, M., Paux, E., Bartoš, J., Feuillet, C.: Chromosome-based genomics in the cereals. — Chromosome Res. 15: 51–66, 2007.
Doležel, J., Lucretti, S.: High-resolution flow karyotyping and chromosome sorting in Vicia faba lines with standard and reconstructed karyotypes. — Theor. appl. Genet. 90: 797–802, 1995.
Doležel, J., Macas, J., Lucretti, S.: Flow analysis and sorting of plant chromosomes. — In: Robinson, J.P., Darzynkiewicz, Z., Dean, P.N., Dressler, L.G., Orfao, A., Rabinovitch, P.S., Stewart, C.C., Tanke, H.J., Wheeless, L.L., (ed.): Current Protocols in Cytometry. Pp. 5.3.1.–5.3.33. John Wiley & Sons, New York 1999.
Doležel, J., Vrána, J., Šafář, J., Bartoš, J., Kubaláková, M., Šimková, H.: Chromosomes in the flow to simplify genome analysis. — Funct. Int. Genom. 12: 397–416, 2012.
Feuillet, C., Eversole, K.: Physical mapping of the wheat genome: A coordinated effort to lay the foundation for genome sequencing and develop tools for breeders. — Isr. J. Plant Sci. 55: 307–313, 2007.
Feuillet, C., Travella, S., Stein, N., Albar, L., Nublat, A., Keller, B.: Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. — Proc. nat. Acad. Sci. USA 100: 15253–15258, 2003.
Gill, K.S., Arumuganathan, K., Le, J.H.: Isolating individual wheat (Triticum aestivum) chromosome arm by flow cytometric analysis of ditelosomic lines. — Theor. appl. Genet. 98: 1248–1252, 1999.
Giorgi, D., Farina, A., Grosso, V., Gennaro, A., Ceoloni, C., Lucretti, S.: FISHIS: fluorescence in situ hybridization in suspension and chromosome flow sorting made easy. — Plos One 8: e57994, 2013.
Hernandez, P., Martis, M., Dorado, G., Pfeifer, M., Gálvez, S., Schaaf, S., Jouve, N., Šimková, H., Valárik, M., Doležel, J., Mayer, K.F.X.: Next-generation sequencing and syntenic integration of flow-sorted arms of wheat chromosome 4A exposes the chromosome structure and gene content. — Plant J. 69: 377–386, 2012.
Huang, L., Brooks, S.A., Li, W.L., Fellers, J.P., Trick, H.N., Gill, B.S.: Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. — Genetics 164: 655–664, 2003.
Jakobson, I., Reis, D., Tiidema, A., Peusha, H., Timofejeva, L., Valárik, M., Kladivová, M., Šimková, H., Doležel, J., Järve, K.: Fine mapping, phenotypic characterization and validation of non-race-specific resistance to powdery mildew in a wheat-Triticum militinae introgression line. — Theor. appl. Genet. 125: 609–623, 2012.
Kantar, M., Akpınar, B.A., Valárik, M., Lucas, S.J., Doležel, J., Hernández, P., Budak, H.: Subgenomic analysis of microRNAs in polyploid wheat. — Funct. int. Genom. 12: 465–479, 2012.
Kejnovský, E., Vrána, J., Matsunaga, S., Souček, P., Široký, J., Doležel, J., Vyskot, B.: Localization of male-specifically expressed MROS genes on Silene latifolia by PCR on flowsorted sex chromosomes and autosomes. — Genetics 158: 1269–1277, 2001.
Kubaláková, M., Kovářová, P., Suchánková, P., Číhalíková, J., Bartoš, J., Lucretti, S., Watanabe, N., Kianian, S.F., Doležel, J.: Chromosome sorting in tetraploid wheat and its potential for genome analysis. — Genetics 170: 823–829, 2005.
Kubaláková, M., Valárik, M., Bartoš, J., Vrána, J., Číhalíková, J., Molnár-Láng, M., Doležel, J.: Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. — Genome 46: 893–905, 2003.
Kubaláková, M., Vrána, J., Čihalíková, J., Šimková, H., Doležel, J.: Flow karyotyping and chromosome sorting in bread wheat (Triticum aestivum L.). — Theor. appl. Genet. 104: 1362–1372, 2002.
Langlois, R.G., Yu, L.-C., Gray, J.W., Carrano, A.V.: Quantitative karyotyping of human chromosomes by dual beam flow cytometry. — Proc. nat. Acad. Sci. USA 79: 7876–7880, 1982.
Lee, J.H., Arumuganathan, K., Yen, Y., Kaeppler, S., Kaeppler, H., Baenziger, P.S.: Root tip cell cycle synchronization and metaphase-chromosome isolation suitable for flow sorting in common wheat (Triticum aestivum L.). — Genome 40: 633–638, 1997.
Li, I.J., Arumuganathan, K., Rines, H.W., Phillips, R.L., Riera-Lizarazu, O., Sandhu, D., Zhou, Y., Gill, K.S.: Flow cytometric sorting of maize chromosome 9 from an oat- maize chromosome addition line. — Theor. appl. Genet. 102: 658–663, 2001.
Lucretti, S., Doležel, J.: Bivariate flow karyotyping in broad bean (Vicia faba). — Cytometry 28: 236–242, 1997.
Ma, L., Xiao, Y., Huang, H., Wang, Q., Rao, W., Feng, Y., Zhang, K., Song, Q.: Direct determination of molecular haplotypes by chromosome microdissection. — Natur. Meth. 7: 299–301, 2010.
Ma, Y.Z., Lee, J.H., Li, L.C., Uchiyama, S., Ohmido, N., Fukui, K.: Fluorescent labeling of plant chromosomes in suspension by FISH. — Gene Genet. Syst. 80: 35–39, 2005.
Macas, J., Doležel, J., Gualberti, G., Pich, U., Schubert, I., Lucretti, S.: Primer-induced labelling of pea and field bean chromosomes in situ and in suspension. — Biotechniques 19: 402–408, 1995.
Madlung, A.: Polyploidy and its effect on evolutionary success: old questions revisited with new tools. — Heredity 110: 99–104, 2013.
Martis, M.M., Klemme, S., Banaei-Moghaddam, A.M., Blattner, F.R., Macas, J., Schmutzer, T., Scholz, U., Gundlach, H., Wicker, T., Šimková, H., Novák, P., Neumann, P., Kubaláková, M., Bauer, E., Haseneyer, G., Fuchs, J., Doležel, J., Stein, N., Mayer, K.F.X., Houben, A.: Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. — Proc. nat. Acad. Sci. USA 109: 13343–13346, 2012.
Mayer, K.F.X., Martis, M., Hedley, P.E., Šimková, H., Liu, H., Morris, J.A., Steuernagel, B., Taudien, S., Roessner, S., Gundlach, H., Kubaláková, M., Suchánková, P., Murat, F., Felder, M., Nussbaumer, T., Graner, A., Salse, J., Endo, T., Sakai, H., Tanaka, T., Itoh, T., Sato, K., Platzer, M.. Matsumoto, T., Scholz, U., Doležel, J., Waugh, R., Stein, N.: Unlocking the barley genome by chromosomal and comparative genomics. — Plant Cell 23: 1249–1263, 2011.
Neumann, P., Lysák, M., Doležel, J., Macas, J.: Isolation of chromosomes from Pisum sativum L. hairy root cultures and their analysis by flow cytometry. — Plant Sci. 137: 205–215, 1998.
Neumann, P., Požárková, D., Vrána, J., Doležel, J., Macas, J.: Chromosome sorting and PCR-based physical mapping in pea (Pisum sativum L.). — Chrom. Res. 10: 63–67, 2002.
Ng, S.B., Turner, E.H., Robertson, P.D., Flygare, S.D., Bigham, A.W., Lee, C., Shaffer, T., Wong, M., Bhattacharjee, A., Eichler, E.E., Bamshad, M., Nickerson, D.A., Shendure, J.: Targeted capture and massively parallel sequencing of 12 human exomes. — Nature 461: 272–278, 2009.
Paterson, A.H.: Leafing through the genomes of our major crop plants: strategies for capturing unique information. — Nat. Rev. Genet. 7: 174–184, 2006.
Paux, E., Sourdille, P., Salse, J., Saintenac, C., Choulet, F., Leroy, P., Korol, A., Michalak, M., Kianian, S., Spielmeyer, W., Lagudah, E., Somers, D., Kilian, A., Alaux, M., Vautrin, S., Berges, H., Eversole, K., Appels, R., Šafář, J., Šimková, H., Doležel, J., Bernard, M., Feuillet, C.: A physical map of the 1-gigabase bread wheat chromosome 3B. — Science 322: 101–104, 2008.
Petrovská, B., Jeřábková, H., Chamrád, I., Vrána, J., Lenobel, R., Uřinovská, J., Šebela, M., Doležel, J.: Proteomic analysis of barley cell nuclei purified by flow sorting. — Cytogenet. Genome Res. 143: 78–86, 2014.
Pich, U., Meister, A., Macas, J., Doležel, J., Lucretti, S., Schubert, I.: Primed in situ labelling facilitates flow sorting of similar sized chromosomes. — Plant J. 7: 1039–1044, 1995.
Požárková, D., Koblížková, A., Román, B., Torres, A.M., Lucretti, S., Lysák, M., Doležel, J., Macas, J.: Development and characterization of microsatellite markers from chromosome 1-specific DNA libraries of Vicia faba. — Biol. Plant. 45: 337–345, 2002.
Šafář, J., Bartoš, J., Janda, J., Bellec, A., Kubaláková, M., Valárik, M., Pateyron, S., Weiserová, J., Tušková, R., Číhalíková, J., Vrána, J., Šimková, H., Faivre-Rampant, P., Sourdille, P., Caboche, M., Bernard, M., Doležel, J., Chalhoub, B.: Dissecting large and complex genomes: flow sorting and BAC cloning of individual chromosomes from bread wheat. — Plant J. 39: 960–968, 2004.
Šafář, J., Šimková, H., Kubaláková, M., Číhalíková, J., Suchánková, P., Bartoš, J., Doležel, J.: Development of chromosome-specific BAC resources for genomics of bread wheat. — Cytogenet. Genome Res. 129: 211–223, 2010.
Sears, E.R., Sears, L.M.S.: The telocentric chromosomes of common wheat. — In: Ramanujam, S. (ed.): Proc. 5th International Wheat Genet Symposium. Pp. 389–407. Indian Society of Genetics and Plant Breeding, IARI, New Delhi 1978.
Shatalina, M., Wicker, T., Buchmann, J.P., Oberhaensli, S., Šimková, H., Doležel, J., Keller, B.: Genotype-specific SNP map based on whole chromosome 3B sequence information from wheat cultivars Arina and Forno. — Plant Biotechnol. J. 11: 23–32, 2013.
Šimková, H., Svensson, J.T., Condamine, P., Hřibová, E., Suchánková, P., Bhat, P.R., Bartoš, J., Šafář, J., Close, T.J., Doležel, J.: Coupling amplified DNA from flow-sorted chromosomes to high-density SNP mapping in barley. — BMC Genomics 9: 237, 2008.
Stein, N., Feuillet, C., Wicker, T., Schlagenhauf, E., Keller, B.: Subgenome chromosome walking in wheat: a 450-kb physical contig in Triticum monococcum L. spans the Lr10 resistance locus in hexaploid wheat (Triticum aestivum L.). — Proc. nat. Acad. Sci. USA 97: 13436–13441, 2000.
Suchánková, P., Kubaláková, M., Kovářová, P., Bartoš, J., Číhalíková, J., Molnár-Láng, M., Endo, T.R., Doležel, J.: Dissection of the nuclear genome of barley by chromosome flow sorting. — Theor. appl. Genet. 113: 651–659, 2006.
Uauy, C., Brevis, J.C., Dubcovsky, J.: The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. — J. exp. Bot. 57: 2785–2794, 2006.
Valárik, M., Bartoš, J., Kovářová, P., Kubaláková, M., De Jong, H., Doležel, J.: High-resolution FISH on super-stretched flow-sorted plant chromosomes. — Plant J. 37: 940–950, 2004.
Van de Peer, Y., Fawcett, J.A., Proost, S., Sterck, L., Vandepoele, K.: The flowering world: a tale of duplications. — Trends Plant Sci. 14: 680–688, 2009.
Vláčilová, K., Ohri, D., Vrána, J., Číhalíková, J., Kubaláková, M., Kahl, G., Doležel, J.: Development of flow cytogenetics and physical genome mapping in chickpea (Cicer arietinum L.). — Chrom. Res. 10: 695–706, 2002.
Vrána, J., Kubaláková, M., Šimková, H., Číhalíková, J., Lysák, M.A., Doležel J.: Flow-sorting of mitotic chromosomes in common wheat (Triticum aestivum L.). — Genetics1 56: 2033–2041, 2000.
Vrána, J., Šimková, H., Kubaláková, M., Číhalíková, J., Doležel, J.: Flow cytometric chromosome sorting in plants: the next generation. — Methods 57: 331–337, 2012.
Wenzl, P., Suchánková, P., Carling, J., Šimková, H., Huttner, E., Kubaláková, M., Sourdille, P., Paul, E., Feuillet, C., Kilian, A., Doležel, J.: Isolated chromosomes as a new and efficient source of DArT markers for the saturation of genetic maps. — Theor. appl. Genet. 121: 465–474, 2010.
Yang, H., Chen, X., Wong, W.H.: Completely phased genome sequencing through chromosome sorting. — Proc. nat. Acad. Sci. USA 108: 12–17, 2011.
Zatloukalová, P., Hřibová, E., Kubaláková, M., Suchánková, P., Šimková, H., Adoración, C., Kahl, G., Millán, T., Doležel, J.: Integration of genetic and physical maps of the chickpea (Cicer arietinum L.) genome using flow-sorted chromosomes. — Chromosome Res. 19: 729–739, 2011.
Zhou, S., Bechner, M.C., Place, M., Churas, C.P., Pape, L., Leong, S.A., Runnheim, R., Forrest, D.K., Goldstein, S., Livny, M., Schwartz, D.C.: Validation of rice genome sequence by optical mapping. — BMC Genom. 8: 278, 2007.
Author information
Authors and Affiliations
Corresponding author
Additional information
Acknowledgements: We warmly acknowledge the help provided by R. Šperková and Z. Dubská in chromosome sorting. We are grateful to P. Sourdille (INRA, Clermont-Ferrand, France) for providing seeds of bread wheat cv. Chinese Spring and Renan, K. Järve (the Tallinn Technical University, Tallinn, Estonia) for seeds of cv. Tähti, and B.S. Gill (the Kansas State University, Manhattan, USA) for seeds of double ditelosomic line 4A of cv. Chinese Spring. This work was supported by a grant award LO1204 from the National Program of Sustainability I.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Vrána, J., Kubaláková, M., Číhalíková, J. et al. Preparation of sub-genomic fractions enriched for particular chromosomes in polyploid wheat. Biol Plant 59, 445–455 (2015). https://doi.org/10.1007/s10535-015-0522-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10535-015-0522-1