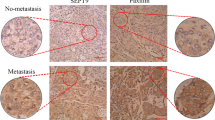
Abstract
Metastasis remains a significant challenge in treating cancer. A better understanding of the molecular mechanisms underlying metastasis is needed to develop more effective treatments. Here, we show that human breast tumor biomarker miR-30c regulates invasion by targeting the cytoskeleton network genes encoding twinfilin 1 (TWF1) and vimentin (VIM). Both VIM and TWF1 have been shown to regulate epithelial-to-mesenchymal transition. Similar to TWF1, VIM also regulates F-actin formation, a key component of cellular transition to a more invasive mesenchymal phenotype. To further characterize the role of the TWF1 pathway in breast cancer, we found that IL-11 is an important target of TWF1 that regulates breast cancer cell invasion and STAT3 phosphorylation. The miR-30c-VIM/TWF1 signaling cascade is also associated with clinical outcome in breast cancer patients.




Similar content being viewed by others
References
Sporn MB (1996) The war on cancer. Lancet 347(9012):1377–1381
Prat A, Perou CM (2011) Deconstructing the molecular portraits of breast cancer. Mol Oncol 5(1):5–23. doi:10.1016/j.molonc.2010.11.003
Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM (2010) Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 12(5):R68. doi:10.1186/bcr2635
Prat A, Perou CM (2009) Mammary development meets cancer genomics. Nat Med 15(8):842–844. doi:10.1038/nm0809-842
Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, Feleppa F, Huschtscha LI, Thorne HJ, Fox SB, Yan M, French JD, Brown MA, Smyth GK, Visvader JE, Lindeman GJ (2009) Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15(8):907–913. doi:10.1038/nm.2000
Bockhorn J, Dalton R, Nwachukwu C, Huang S, Prat A, Yee K, Chang Y-F, Huo D, Wen Y, Swanson KE, Qiu T, Lu J, Park SY, Dolan ME, Perou CM, Olopade OI, Clarke MF, Greene GL, Liu H (2012) MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nature Commun (in press)
Yang J, Weinberg RA (2008) Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14(6):818–829. doi:10.1016/j.devcel.2008.05.009
Singh A, Settleman J (2010) EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29(34):4741–4751. doi:10.1016/j.devcel.2008.05.009
Meacham CE, Ho EE, Dubrovsky E, Gertler FB, Hemann MT (2009) In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nat Genet 41(10):1133–1137
Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, Wen S, Chang YF, Bachmann MH, Shimono Y, Dalerba P, Adorno M, Lobo N, Bueno J, Dirbas FM, Goswami S, Somlo G, Condeelis J, Contag CH, Gambhir SS, Clarke MF (2010) Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A 107(42):18115–18120
Team RDC (2011) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, Taylor M, Harris AL, Ragoussis J (2011) microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res 71(17):5635–5645
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65(16):7065–7070. doi:10.1158/0008-5472.CAN-05-1783
Liu H (2012) MicroRNAs in breast cancer initiation and progression. Cell Mol Life Sci. doi:10.1007/s00018-012-1128-9
Antonov AV, Dietmann S, Wong P, Lutter D, Mewes HW (2009) GeneSet2miRNA: finding the signature of cooperative miRNA activities in the gene lists. Nucleic Acids Res 37 (Web Server issue):W323-328. doi: 10.1093/nar/gkp313
Palmgren S, Vartiainen M, Lappalainen P (2002) Twinfilin, a molecular mailman for actin monomers. J Cell Sci 115(Pt 5):881–886
Ojala PJ, Paavilainen VO, Vartiainen MK, Tuma R, Weeds AG, Lappalainen P (2002) The two ADF-H domains of twinfilin play functionally distinct roles in interactions with actin monomers. Mol Biol Cell 13(11):3811–3821. doi:10.1091/mbc.E02-03-0157
Poukkula M, Kremneva E, Serlachius M, Lappalainen P (2011) Actin-depolymerizing factor homology domain: a conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton (Hoboken) 68(9):471–490. doi:10.1002/cm.20530
Ernst M, Najdovska M, Grail D, Lundgren-May T, Buchert M, Tye H, Matthews VB, Armes J, Bhathal PS, Hughes NR, Marcusson EG, Karras JG, Na S, Sedgwick JD, Hertzog PJ, Jenkins BJ (2008) STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J Clin Invest 118(5):1727–1738. doi:10.1172/JCI34944
Schuringa JJ, Wierenga AT, Kruijer W, Vellenga E (2000) Constitutive Stat3, Tyr705, and Ser727 phosphorylation in acute myeloid leukemia cells caused by the autocrine secretion of interleukin-6. Blood 95(12):3765–3770
Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA-target recognition. PLoS Biol 3(3):e85. doi:10.1371/journal.pbio.0030085
Li N, Kaur S, Greshock J, Lassus H, Zhong X, Wang Y, Leminen A, Shao Z, Hu X, Liang S, Katsaros D, Huang Q, Butzow R, Weber BL, Coukos G, Zhang L (2012) A combined array-based comparative genomic hybridization and functional library screening approach identifies mir-30d as an oncomir in cancer. Cancer Res 72(1):154–164. doi:10.1158/0008-5472.CAN-11-2484
Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, de Vega-Saenz Miera E, Rakus JF, Dankert JF, Shang S, Kerbel RS, Bhardwaj N, Shao Y, Darvishian F, Zavadil J, Erlebacher A, Mahal LK, Osman I, Hernando E (2011) miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 20(1):104–118. doi:10.1016/j.ccr.2011.05.027
Elson-Schwab I, Lorentzen A, Marshall CJ (2010) MicroRNA-200 family members differentially regulate morphological plasticity and mode of melanoma cell invasion. PloS one 5 (10). doi:10.1371/journal.pone.0013176
Cheng CW, Wang HW, Chang CW, Chu HW, Chen CY, Yu JC, Chao JI, Liu HF, Ding SL, Shen CY (2012) MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res Treat 134(3):1081–1093. doi:10.1007/s10549-012-2034-4
Acknowledgments
We are very thankful to Dr. Jun Lu at Yale University for providing the miR-30c precursor gene vector, Dr. Seo Young Park for data analyses, and Dr. John Kokontis for helping review and edit our manuscript. We appreciate the experimental support of several core facilities, including the Animal Facility, Optical Imaging Core Facility, Flow Cytometry Facility, Integrated Microscopy Core Facility, DNA Sequencing Facility, Functional Genomics Facility, and IHC Core Facility at the University of Chicago. We specifically acknowledge Ryan Duggan, James Cao, David Leclerc, Marianne Greene, Terri Li, Xin Jiang, Shirley Bond, Jaejung Kim, Hui Zheng, Andrew Gusev, Dalong Qian, and Yohei Shimono for technical help and support.
Funding
This study was supported in part by The University of Chicago Women’s Board (J.B.), National Institutes of Health (NIH) T90 Regenerative Medicine Training Program DK070103-05, Department of Defense Breast Cancer Research Program W81XWH-09-1-0331, Paul Calabresi K12 Award 1K12CA139160-02, NCI K99 CA160638-01A1, Chicago Fellows Program at the University of Chicago, and the University of Chicago Clinical and Translational Science Award (UL1 RR024999) (H.L.), funds from the Sociedad Española de Oncología Médica (SEOM) (A.P.), the Breast SPORE at University of North Carolina 5-P50-CA58223-17 (A.P. and C.M.P.), the Breast SPORE at the University of Chicago P50CA125183-05, the Doris Duke Charitable Foundation (O.I.O and C.N.), The University of Chicago Cancer Research Center Pilot Fund, BSD Imaging Research Institute Pilot Research Projects Using Animal Imaging, UCMC/Northshore Collaborative Funds, a Carole and Gordon Segal Grant (G.L.G.), and the Virginia and D. K. Ludwig Fund (G.L.G. and H.L.), NIH Grants U54 CA 126524 and P01 CA139490 (M.F.C.), the Breast Cancer Research Foundation (M.F.C., O.I.O. and C.M.P), and the University of Chicago Cancer Center Support Grant CA 014599.
Conflicts of interest
Authors of this manuscript have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bockhorn, J., Yee, K., Chang, YF. et al. MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Cancer Res Treat 137, 373–382 (2013). https://doi.org/10.1007/s10549-012-2346-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-012-2346-4