Abstract
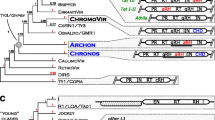
Long terminal repeat (LTR) retrotransposons make up substantial parts of most higher plant genomes where they accumulate due to their replicative mode of transposition. Although the transposition is facilitated by proteins encoded within the gag-pol region which is common to all autonomous elements, some LTR retrotransposons were found to potentially carry an additional protein coding capacity represented by extra open reading frames located upstream or downstream of gag-pol. In this study, we performed a comprehensive in silico survey and comparative analysis of these extra open reading frames (ORFs) in the group of Ty3/gypsy LTR retrotransposons as the first step towards our understanding of their origin and function. We found that extra ORFs occur in all three major lineages of plant Ty3/gypsy elements, being the most frequent in the Tat lineage where most (77 %) of identified elements contained extra ORFs. This lineage was also characterized by the highest diversity of extra ORF arrangement (position and orientation) within the elements. On the other hand, all of these ORFs could be classified into only two broad groups based on their mutual similarities or the presence of short conserved motifs in their inferred protein sequences. In the Athila lineage, the extra ORFs were confined to the element 3′ regions but they displayed much higher sequence diversity compared to those found in Tat. In the lineage of Chromoviruses the extra ORFs were relatively rare, occurring only in 5′ regions of a group of elements present in a single plant family (Poaceae). In all three lineages, most extra ORFs lacked sequence similarities to characterized gene sequences or functional protein domains, except for two Athila-like elements with similarities to LOGL4 gene and part of the Chromoviruses extra ORFs that displayed partial similarity to histone H3 gene. Thus, in these cases the extra ORFs most likely originated by transduction or recombination of cellular gene sequences. In addition, the protein domain which is otherwise associated with DNA transposons have been detected in part of the Tat-like extra ORFs, pointing to their origin from an insertion event of a mobile element.






Similar content being viewed by others
References
Babu MM, Iyer LM, Balaji S, Aravind L (2006) The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res 34:6505–6520
Barbeau B, Mesnard J-M (2011) Making sense out of antisense transcription in human T-cell lymphotropic viruses (HTLVs). Viruses 3:456–468
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Coffin JM, Hughes SH, Varmus HE (1997) Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Du J, Tian Z, Hans CS, Laten HM, Cannon SB, Jackson SA, Shoemaker RC, Ma J (2010) Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J 63:584–598
Elrouby N, Bureau TE (2001) A novel hybrid open reading frame formed by multiple cellular gene transductions by a plant long terminal repeat retroelement. J Biol Chem 276:41963–41968
Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A (2010) The Pfam protein families database. Nucleic Acids Res 38:D211–D222
Forbes EM, Nieduszynska SR, Brunton FK, Gibson J, Glover LA, Stansfield I (2007) Control of gag-pol gene expression in the Candida albicans retrotransposon Tca2. BMC Mol Biol 8:94
Gao X, Havecker ER, Baranov PV, Atkins JF, Voytas DF (2003) Translational recoding signals between gag and pol in diverse LTR retrotransposons. RNA 9:1422–1430
Gao D, Gill N, Kim H-R, Walling JG, Zhang W, Fan C, Yu Y, Ma J, SanMiguel P, Jiang N, Cheng Z, Wing RA, Jiang J, Jackson SA (2009) A lineage-specific centromere retrotransposon in Oryza brachyantha. Plant J 60:820–831
Gorinsek B, Gubensek F, Kordis D (2004) Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol 21:781–798
Havecker ER, Gao X, Voytas DF (2004) The diversity of LTR retrotransposons. Genome Biol 5:225
Hawkins JS, Grover CE, Wendel JF (2008) Repeated big bangs and the expanding universe: directionality in plant genome size evolution. Plant Sci 174:557–562
Hofmann K, Stoffel W (1993) TMBASE—a database of membrane spanning protein segments. Biol Chem H-S 374:166
Hu TT, Pattyn P, Bakker EG et al (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43:476–481
International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768
Jaillon O, Aury J-M, Noel B et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jin YK, Bennetzen JL (1994) Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell 6:1177–1186
Kato A, Endo M, Kato H, Saito T (2005) The antisense promoter of AtRE1, a retrotransposon in Arabidopsis thaliana, is activated in pollens and calluses. Plant Sci 168:981–986
Kejnovsky E, Kubat Z, Macas J, Hobza R, Mracek J, Vyskot B (2006) Retand: a novel family of gypsy-like retrotransposons harboring an amplified tandem repeat. Mol Genet Genomics 76:254–263
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Kumekawa N, Ohtsubo H, Horiuchi T, Ohtsubo E (1999) Identification and characterization of novel retrotransposons of the gypsy type in rice. Mol Gen Genet 260:593–602
Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H (2009) Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21:3152–3169
Laten HM, Mogil LS, Wright LN (2009) A shotgun approach to discovering and reconstructing consensus retrotransposons ex novo from dense contigs of short sequences derived from Genbank Genome Survey Sequence database records. Gene 448:168–173
Li W, Jaroszewski L, Godzik A (2001) Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 17:282–283
Li W, Jaroszewski L, Godzik A (2002) Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 18:77–82
Lloréns C, Futami R, Bezemer D, Moya A (2008) The gypsy database (GyDB) of mobile genetic elements. Nucleic Acids Res 36:D38–D46
Loidl P (2004) A plant dialect of the histone language. Trends Plant Sci 9:84–90
Macas J, Neumann P (2007) Ogre elements—a distinct group of plant Ty3/gypsy-like retrotransposons. Gene 390:108–116
Macas J, Koblížková A, Navrátilová A, Neumann P (2009) Hypervariable 3′ UTR region of plant LTR-retrotransposons as a source of novel satellite repeats. Gene 448:198–206
Macas J, Kejnovský E, Neumann P, Novák P, Koblížková A, Vyskot B (2011) Next generation sequencing-based analysis of repetitive DNA in the model dioecious plant Silene latifolia. PLoS ONE 6:e27335
Marchler-Bauer A, Lu S, Anderson JB et al (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229
Marín I, Lloréns C (2000) Ty3/Gypsy retrotransposons: description of new Arabidopsis thaliana elements and evolutionary perspectives derived from comparative genomic data. Mol Biol Evol 17:1040–1049
Martínez-Izquierdo JA, García-Martínez J, Vicient CM (1997) What makes Grande1 retrotransposon different? Genetica 100:15–28
McCarthy EM, Liu J, Lizhi G, McDonald JF (2002) Long terminal repeat retrotransposons of Oryza sativa. Genome Biol 3 (RESEARCH0053)
Ming R, Hou S, Feng Y et al (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452:991–996
Neumann P, Požárková D, Macas J (2003) Highly abundant pea LTR retrotransposon Ogre is constitutively transcribed and partially spliced. Plant Mol Biol 53:399–410
Neumann P, Požárková D, Koblížková A, Macas J (2005) PIGY, a new plant envelope-class LTR retrotransposon. Mol Genet Genomics 273:43–53
Neumann P, Koblížková A, Navrátilová A, Macas J (2006) Significant expansion of Vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics 173:1047–1056
Neumann P, Navrátilová A, Koblížková A, Kejnovský E, Hřibová E, Hobza R, Widmer A, Doležel J, Macas J (2011) Plant centromeric retrotransposons: a structural and cytogenetic perspective. Mobile DNA 2:4
Novák P, Neumann P, Macas J (2010) Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinform 11:378
Ohtsubo H, Kumekawa N, Ohtsubo E (1999) RIRE2, a novel gypsy-type retrotransposon from rice. Genes Genet Syst 74:83–91
Ouyang S, Zhu W, Hamilton J et al (2007) The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35:D883–D887
Paterson AH, Bowers JE, Bruggmann R et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448
Pearson WR, Wood T, Zhang Z, Miller W (1997) Comparison of DNA sequences with protein sequences. Genomics 46:24–36
Pereira V (2004) Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol 5:R79
Peterson-Burch BD, Wright DA, Laten HM, Voytas DF (2000) Retroviruses in plants? Trends Genet 16:151–152
Schmutz J, Cannon SB, Schlueter J et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Steinbauerová V, Neumann P, Macas J (2008) Experimental evidence for splicing of intron-containing transcripts of plant LTR retrotransposon Ogre. Mol Genet Genomics 280:427–436
Tuskan GA, Difazio S, Jansson S et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Vicient CM, Kalendar R, Schulman AH (2001) Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res 11:2041–2049
Wicker T, Keller B (2007) Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res 17:1072–1081
Wright DA, Voytas DF (2002) Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res 12:122–131
Xu Z, Wang H (2007) LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35:W265–W268
Yano ST, Panbehi B, Das A, Laten HM (2005) Diaspora, a large family of Ty3-gypsy retrotransposons in Glycine max, is an envelope-less member of an endogenous plant retrovirus lineage. BMC Evol Biol 5:30
Zuccolo A, Sebastian A, Talag J, Yu Y, Kim H, Collura K, Kudrna D, Wing RA (2007) Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evol Biol 7:152
Acknowledgments
We thank Jasper E. Manning for his help with manuscript preparation. This work was supported by grants AVOZ50510513 from the Academy of Sciences of the Czech Republic, and P501/12/G090 from the Czech Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Steinbauerová, V., Neumann, P., Novák, P. et al. A widespread occurrence of extra open reading frames in plant Ty3/gypsy retrotransposons. Genetica 139, 1543–1555 (2011). https://doi.org/10.1007/s10709-012-9654-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-012-9654-9