Abstract
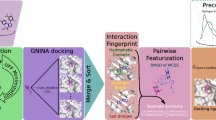
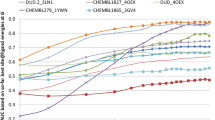
We describe the performance of multiple pose prediction methods for the D3R 2016 Grand Challenge. The pose prediction challenge includes 36 ligands, which represent 4 chemotypes and some miscellaneous structures against the FXR ligand binding domain. In this study we use a mix of fully automated methods as well as human-guided methods with considerations of both the challenge data and publicly available data. The methods include ensemble docking, colony entropy pose prediction, target selection by molecular similarity, molecular dynamics guided pose refinement, and pose selection by visual inspection. We evaluated the success of our predictions by method, chemotype, and relevance of publicly available data. For the overall data set, ensemble docking, visual inspection, and molecular dynamics guided pose prediction performed the best with overall mean RMSDs of 2.4, 2.2, and 2.2 Å respectively. For several individual challenge molecules, the best performing method is evaluated in light of that particular ligand. We also describe the protein, ligand, and public information data preparations that are typical of our binding mode prediction workflow.









Similar content being viewed by others
References
Makishima M, Okamoto AY, Repa JJ et al (1999) Identification of a nuclear receptor for bile acids. Science 284:1362–1365. doi:10.1126/science.284.5418.1362
Goodwin B, Jones SA, Price RR et al (2000) A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 6:517–526. doi:10.1016/S1097-2765(00)00051-4
Feng S, Yang M, Zhang Z et al (2009) Identification of an N-oxide pyridine GW4064 analog as a potent FXR agonist. Bioorg Med Chem Lett 19:2595–2598. doi:10.1016/j.bmcl.2009.03.008
Gao Y, Hu Y, Crespo A et al (2017) Workflows and performances in the ranking prediction of the 2016 Grand Challenge 2: lessons learned from a collaborative effort. J Comput Aided Mol Des (in press)
Walters WP, Stahl MT, Murcko MA (1998) Virtual screening—an overview. Drug Discov Today 3:160–178. doi:10.1016/S1359-6446(97)01163-X
Tanrikulu Y, Krüger B, Proschak E (2013) The holistic integration of virtual screening in drug discovery. Drug Discov Today 18:358–364. doi:10.1016/j.drudis.2013.01.007
Kumar A, Zhang KYJ (2015) Hierarchical virtual screening approaches in small molecule drug discovery. Methods 71:26–37. doi:10.1016/j.ymeth.2014.07.007
Verkhivker GM, Bouzida D, Gehlhaar DK et al (2000) Deciphering common failures in molecular docking of ligand-protein complexes. J Comput Aided Mol Des 14:731–751. doi:10.1023/A:1008158231558
Osterberg F, Morris GM, Sanner MF et al (2002) Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins 46:34–40
Huang S-Y, Zou X (2007) Ensemble docking of multiple protein structures: Considering protein structural variations in molecular docking. Proteins Struct Funct Bioinform 66:399–421. doi:10.1002/prot.21214
Ellingson SR, Miao Y, Baudry J, Smith JC (2015) Multi-conformer ensemble docking to difficult protein targets. J Phys Chem B 119:1026–1034. doi:10.1021/jp506511p
Ruvinsky AM, Kozintsev AV (2006) Novel statistical-thermodynamic methods to predict protein-ligand binding positions using probability distribution functions. Proteins 62:202–208. doi:10.1002/prot.20673
Ruvinsky AM, Kozintsev AV (2005) New and fast statistical-thermodynamic method for computation of protein-ligand binding entropy substantially improves docking accuracy. J Comput Chem 26:1089–1095. doi:10.1002/jcc.20246
Ruvinsky AM (2007) Role of binding entropy in the refinement of protein-ligand docking predictions: analysis based on the use of 11 scoring functions. J Comput Chem 28:1364–1372. doi: 10.1002/jcc.20580
Friesner RA, Banks JL, Murphy RB et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749. doi:10.1021/jm0306430
Jones G, Willett P, Glen RC et al (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748. doi:10.1006/jmbi.1996.0897
Wang R, Lai L, Wang S (2002) Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des 16:11–26
Zhao H, Caflisch A (2015) Molecular dynamics in drug design. Eur J Med Chem 91:4–14. doi:10.1016/j.ejmech.2014.08.004
Bharatham N, Finch KE, Min J et al (2017) Performance of a docking/molecular dynamics protocol for virtual screening of nutlin-class inhibitors of Mdmx. J Mol Graph Model 74:54–60. doi:10.1016/j.jmgm.2017.02.014
Liu K, Watanabe E, Kokubo H (2017) Exploring the stability of ligand binding modes to proteins by molecular dynamics simulations. J Comput Aided Mol Des 31:201–211. doi:10.1007/s10822-016-0005-2
Berman HM, Westbrook J, Feng Z et al (2000) The protein data bank. Nucleic Acids Res 28:235–242. doi:10.1093/nar/28.1.235
Kleywegt GJ (1996) Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr D Biol Crystallogr 52:842–857. doi:10.1107/S0907444995016477
Sastry GM, Adzhigirey M, Day T et al (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234. doi:10.1007/s10822-013-9644-8
Shelley JC, Cholleti A, Frye LL et al (2007) Epik: a software program for pKa prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des 21:681–691. doi:10.1007/s10822-007-9133-z
Hawkins PCD, Skillman AG, Warren GL et al (2010) Conformer generation with OMEGA: algorithm and validation using high quality structures from the protein databank and cambridge structural database. J Chem Inf Model 50:572–584. doi:10.1021/ci100031x
Hawkins PCD, Skillman AG, Nicholls A (2007) Comparison of shape-matching and docking as virtual screening tools. J Med Chem 50:74–82. doi:10.1021/jm0603365
Case DA, Cheatham TE, Darden T et al (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688. doi: 10.1002/jcc.20290
(2017) Molecular operating environment (MOE). Chemical Computing Group Inc., Montreal
(2016) Pipeline pilot. Dassault Systèmes BIOVIA, San Diego
Wang J, Wang W, Kollman PA, Case DA (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25:247–260. doi:10.1016/j.jmgm.2005.12.005
Sherman W, Day T, Jacobson MP et al (2006) Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem 49:534–553. doi:10.1021/jm050540c
Bowers K, Chow E, Xu H et al (2006) Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. Proc. ACMIEEE Conf. Supercomput. SC06
Verdonk ML, Mortenson PN, Hall RJ et al (2008) Protein—ligand docking against non-native protein conformers. J Chem Inf Model 48:2214–2225. doi:10.1021/ci8002254
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Exp Opin Drug Discov 10:449–461. doi:10.1517/17460441.2015.1032936
Onufriev A, Bashford D, Case DA (2004) Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 394:383–394. doi:10.1002/prot.20033
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Fradera, X., Verras, A., Hu, Y. et al. Performance of multiple docking and refinement methods in the pose prediction D3R prospective Grand Challenge 2016. J Comput Aided Mol Des 32, 113–127 (2018). https://doi.org/10.1007/s10822-017-0053-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-017-0053-2