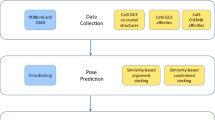
Abstract
During the last few years, we have developed a docking protocol involving two steps: (i) the choice of the most appropriate docking software and parameters for the system of interest using structural and functional information available in public databases (PDB, ChEMBL, PubChem Assay, BindingDB, etc.); (ii) the docking of ligand dataset to provide a prediction for the binding modes and ranking of ligands. We applied this protocol to the D3R Grand Challenge 3 dataset containing cathepsin S (CatS) inhibitors. Considering the size and conformational flexibility of ligands, the docking calculations afforded reasonable overall pose predictions, which are however dependent on the specific nature of each ligand. As expected, the correct ranking of docking poses is still challenging. Post-processing of docking poses with molecular dynamics simulations in explicit solvent provided a significantly better prediction, whereas free energy calculations on a subset of compounds brought no significant improvement in the ranking prediction compared with the direct ranking obtained from the scoring function.








Similar content being viewed by others
References
Thurmond RL, Sun S, Karlsson L, Edwards JP (2005) Cathepsin S inhibitors as novel immunomodulators. Curr Opin Investig Drugs 6(5):473–482
Link JO, Zipfel S (2006) Advances in cathepsin S inhibitor design. Curr Opin Drug Discov Dev 9(4):471–482
Wiener JJ, Sun S, Thurmond RL (2010) Recent advances in the design of cathepsin S inhibitors. Curr Topics Med Chem 10(7):717–732
Lee-Dutra A, Wiener DK, Sun S (2011) Cathepsin S inhibitors: 2004–2010. Expert Opin Ther Pat 21(3):311–337. https://doi.org/10.1517/13543776.2011.553800
Wilkinson RD, Williams R, Scott CJ, Burden RE (2015) Cathepsin S: therapeutic, diagnostic, and prognostic potential. Biol Chem 396(8):867–882. https://doi.org/10.1515/hsz-2015-0114
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28(1):235–242
Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD (2003) Improved protein–ligand docking using GOLD. Proteins Struct Funct Bioinf 52(4):609–623. https://doi.org/10.1002/prot.10465
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
Selwa E, Elisée E, Zavala A, Iorga BI (2018) Blinded evaluation of farnesoid X receptor (FXR) ligands binding using molecular docking and free energy calculations. J Comput Aided Mol Des 32(1):273–286. https://doi.org/10.1007/s10822-017-0054-1
Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29(7):845–854. https://doi.org/10.1093/bioinformatics/btt055
Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL (2001) Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B 105(28):6474–6487. https://doi.org/10.1021/jp003919d
Robertson MJ, Tirado-Rives J, Jorgensen WL (2015) Improved peptide and protein torsional energetics with the OPLSAA force field. J Chem Theor Comput 11(7):3499–3509. https://doi.org/10.1021/acs.jctc.5b00356
Gapsys V, Michielssens S, Seeliger D, de Groot BL (2015) pmx: automated protein structure and topology generation for alchemical perturbations. J Comput Chem 36(5):348–354. https://doi.org/10.1002/jcc.23804
Gapsys V, Michielssens S, Peters JH, de Groot BL, Leonov H (2015) Calculation of binding free energies. Methods Mol Biol 1215:173–209. https://doi.org/10.1007/978-1-4939-1465-4_9
Gapsys V, Michielssens S, Seeliger D, de Groot BL (2016) Accurate and rigorous prediction of the changes in protein free energies in a large-scale mutation scan. Angew Chem 55(26):7364–7368. https://doi.org/10.1002/anie.201510054
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612. https://doi.org/10.1002/jcc.20084
Surpateanu G, Iorga BI (2012) Evaluation of docking performance in a blinded virtual screening of fragment-like trypsin inhibitors. J Comput Aided Mol Des 26(5):595–601. https://doi.org/10.1007/s10822-011-9526-x
Colas C, Iorga BI (2014) Virtual screening of the SAMPL4 blinded HIV integrase inhibitors dataset. J Comput Aided Mol Des 28(4):455–462. https://doi.org/10.1007/s10822-014-9707-5
Martiny VY, Martz F, Selwa E, Iorga BI (2016) Blind pose prediction, scoring, and affinity ranking of the CSAR 2014 dataset. J Chem Inform Model 56(6):996–1003. https://doi.org/10.1021/acs.jcim.5b00337
Selwa E, Martiny VY, Iorga BI (2016) Molecular docking performance evaluated on the D3R Grand Challenge 2015 drug-like ligand datasets. J Comput Aided Mol Des 30(9):829–839. https://doi.org/10.1007/s10822-016-9983-3
Acknowledgements
We thank Prof. Bert de Groot for helpful discussions. This work was supported by the Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT) (Agence Nationale de la Recherche, Grant Number ANR-10-LABX-33), by the JPIAMR transnational project DesInMBL (Agence Nationale de la Recherche, Grant Number ANR-14-JAMR-0002) and by the Région Ile-de-France (Conseil Régional, Île-de-France, DIM Malinf).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10822_2018_161_MOESM1_ESM.pdf
The Electronic Supplementary Material contains the list of CatS crystal structures from the PDB, the chemical structures of the scoring CatS D3RGC3 dataset, and the plots showing the performance of our submissions.
Rights and permissions
About this article
Cite this article
Chaput, L., Selwa, E., Elisée, E. et al. Blinded evaluation of cathepsin S inhibitors from the D3RGC3 dataset using molecular docking and free energy calculations. J Comput Aided Mol Des 33, 93–103 (2019). https://doi.org/10.1007/s10822-018-0161-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0161-7