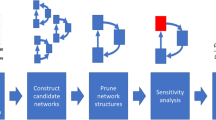
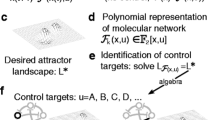
Abstract
Quantitative systems pharmacology (QSP) is an emerging discipline that aims to discover how drugs modulate the dynamics of biological components in molecular and cellular networks and the impact of those perturbations on human pathophysiology. The integration of systems-based experimental and computational approaches is required to facilitate the advancement of this field. QSP models typically consist of a series of ordinary differential equations (ODE). However, this mathematical framework requires extensive knowledge of parameters pertaining to biological processes, which is often unavailable. An alternative framework that does not require knowledge of system-specific parameters, such as Boolean network modeling, could serve as an initial foundation prior to the development of an ODE-based model. Boolean network models have been shown to efficiently describe, in a qualitative manner, the complex behavior of signal transduction and gene/protein regulatory processes. In addition to providing a starting point prior to quantitative modeling, Boolean network models can also be utilized to discover novel therapeutic targets and combinatorial treatment strategies. Identifying drug targets using a network-based approach could supplement current drug discovery methodologies and help to fill the innovation gap across the pharmaceutical industry. In this review, we discuss the process of developing Boolean network models and the various analyses that can be performed to identify novel drug targets and combinatorial approaches. An example for each of these analyses is provided using a previously developed Boolean network of signaling pathways in multiple myeloma. Selected examples of Boolean network models of human (patho-)physiological systems are also reviewed in brief.

Reproduced with permission from [43] (Color figure online)

Figure reproduced with permission from Chudasama et al. [23] (Color figure online)






Similar content being viewed by others
References
Kitano H (2002) Systems biology: a brief overview. Science 295(5560):1662–1664
Kitano H (2002) Computational systems biology. Nature 420(6912):206–210
Sorger PK, Allerheiligen SR, Abernethy DR, Altman RB, Brouwer KL, Califano A, D’Argenio DZ, Iyengar R, Jusko WJ, Lalonde R Quantitative and systems pharmacology in the post-genomic era: new approaches to discovering drugs and understanding therapeutic mechanisms. In: An NIH white paper by the QSP workshop group, 2011. NIH Bethesda
Iyengar R, Zhao S, Chung S-W, Mager DE, Gallo JM (2012) Merging systems biology with pharmacodynamics. Science translational medicine 4 (126):126ps127
Barabasi A-L, Oltvai ZN (2004) Network biology: understanding the cell’s functional organization. Nat Rev Genet 5(2):101–113
Hwang WC, Zhang A, Ramanathan M (2008) Identification of information flow-modulating drug targets: a novel bridging paradigm for drug discovery. Clin Pharmacol Ther 84(5):563–572
Berger SI, Iyengar R (2009) Network analyses in systems pharmacology. Bioinformatics 25(19):2466–2472
Birtwistle M, Mager D, Gallo J (2013) Mechanistic vs. empirical network models of drug action. CPT 2(9):1–3
Harrold JM, Ramanathan M, Mager DE (2013) Network-based approaches in drug discovery and early development. Clin Pharmacol Ther 94(6):651–658
Kauffman SA (1969) Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol 22(3):437–467
Martínez-Sosa P, Mendoza L (2013) The regulatory network that controls the differentiation of T lymphocytes. Biosystems 113(2):96–103
Saez-Rodriguez J, Simeoni L, Lindquist JA, Hemenway R, Bommhardt U, Arndt B, Haus U-U, Weismantel R, Gilles ED, Klamt S (2007) A logical model provides insights into T cell receptor signaling. PLoS Comput Biol 3(8):e163
Martinez-Sanchez ME, Mendoza L, Villarreal C, Alvarez-Buylla ER (2015) A minimal regulatory network of extrinsic and intrinsic factors recovers observed patterns of CD4 + T cell differentiation and plasticity. PLoS Comput Biol 11(6):e1004324
Mendez A, Mendoza L (2016) A network model to describe the terminal differentiation of B cells. PLoS Comput Biol 12(1):e1004696
Conroy BD, Herek TA, Shew TD, Latner M, Larson JJ, Allen L, Davis PH, Helikar T, Cutucache CE (2014) Design, assessment, and in vivo evaluation of a computational model illustrating the role of CAV1 in CD4 + T-lymphocytes. Frontiers Immunol 5:599
Zhang R, Shah MV, Yang J, Nyland SB, Liu X, Yun JK, Albert R, Loughran TP (2008) Network model of survival signaling in large granular lymphocyte leukemia. Proc Natl Acad Sci 105(42):16308–16313
Saadatpour A, Wang R-S, Liao A, Liu X, Loughran TP, Albert I, Albert R (2011) Dynamical and structural analysis of a T cell survival network identifies novel candidate therapeutic targets for large granular lymphocyte leukemia. PLoS Comput Biol 7(11):e1002267
Oyeyemi OJ, Davies O, Robertson DL, Schwartz JM (2015) A logical model of HIV-1 interactions with the T-cell activation signalling pathway. Bioinformatics 31(7):1075–1083
Rodríguez A, Sosa D, Torres L, Molina B, Frías S, Mendoza L (2012) A Boolean network model of the FA/BRCA pathway. Bioinformatics 28(6):858–866
Rodríguez A, Torres L, Juárez U, Sosa D, Azpeitia E, García-de Teresa B, Cortés E, Ortíz R, Salazar AM, Ostrosky-Wegman P (2015) Fanconi anemia cells with unrepaired DNA damage activate components of the checkpoint recovery process. Theor Biol Med Model 12(1):19
Ruiz-Cerdá ML, Irurzun-Arana I, González-Garcia I, Hu C, Zhou H, Vermeulen A, Trocóniz IF, Gómez-Mantilla JD (2016) Towards patient stratification and treatment in the autoimmune disease lupus erythematosus using a systems pharmacology approach. Eur J Pharm Sci 94:46–58
Walsh ER, Thakar J, Stokes K, Huang F, Albert R, August A (2011) Computational and experimental analysis reveals a requirement for eosinophil-derived IL-13 for the development of allergic airway responses in C57BL/6 mice. J Immunol 186(5):2936–2949
Chudasama VL, Ovacik MA, Abernethy DR, Mager DE (2015) Logic-based and cellular pharmacodynamic modeling of bortezomib responses in U266 human myeloma cells. J Pharmacol Exp Ther 354(3):448–458
Helikar T, Kochi N, Kowal B, Dimri M, Naramura M, Raja SM, Band V, Band H, Rogers JA (2013) A comprehensive, multi-scale dynamical model of ErbB receptor signal transduction in human mammary epithelial cells. PLoS ONE 8(4):e61757
Zhu P, Aliabadi HM, Uludag H, Han J (2016) Identification of potential drug targets in cancer signaling pathways using stochastic logical models. Sci Rep 6:23078
Kirouac DC, Du JY, Lahdenranta J, Overland R, Yarar D, Paragas V, Pace E, McDonagh CF, Nielsen UB, Onsum MD (2013) Computational modeling of ERBB2-amplified breast cancer identifies combined ErbB2/3 blockade as superior to the combination of MEK and AKT inhibitors. Sci Signal 6(288):ra68
von der Heyde S, Bender C, Henjes F, Sonntag J, Korf U, Beissbarth T (2014) Boolean ErbB network reconstructions and perturbation simulations reveal individual drug response in different breast cancer cell lines. BMC Syst Biol 8:75
Sahin O, Frohlich H, Lobke C, Korf U, Burmester S, Majety M, Mattern J, Schupp I, Chaouiya C, Thieffry D, Poustka A, Wiemann S, Beissbarth T, Arlt D (2009) Modeling ERBB receptor-regulated G1/S transition to find novel targets for de novo trastuzumab resistance. BMC Syst Biol 3:1
Choi M, Shi J, Jung SH, Chen X, Cho KH (2012) Attractor landscape analysis reveals feedback loops in the p53 network that control the cellular response to DNA damage. Sci Signal 5(251):ra83
Flobak A, Baudot A, Remy E, Thommesen L, Thieffry D, Kuiper M, Laegreid A (2015) Discovery of drug synergies in gastric cancer cells predicted by logical modeling. PLoS Comput Biol 11(8):e1004426
Trairatphisan P, Wiesinger M, Bahlawane C, Haan S, Sauter T (2016) A probabilistic Boolean network approach for the analysis of cancer-specific signalling: a case study of deregulated PDGF signalling in GIST. PLoS ONE 11(5):e0156223
Lu J, Zeng H, Liang Z, Chen L, Zhang L, Zhang H, Liu H, Jiang H, Shen B, Huang M, Geng M, Spiegel S, Luo C (2015) Network modelling reveals the mechanism underlying colitis-associated colon cancer and identifies novel combinatorial anti-cancer targets. Sci Rep 5:14739
Saez-Rodriguez J, Alexopoulos LG, Zhang MS, Morris MK, Lauffenburger DA, Sorger PK (2011) Comparing signaling networks between normal and transformed hepatocytes using discrete logical models. Cancer Res 71(16):5400–5411
Steinway SN, Zanudo JG, Ding W, Rountree CB, Feith DJ, Loughran TP Jr, Albert R (2014) Network modeling of TGFbeta signaling in hepatocellular carcinoma epithelial-to-mesenchymal transition reveals joint sonic hedgehog and Wnt pathway activation. Cancer Res 74(21):5963–5977
Udyavar AR, Wooten DJ, Hoeksema M, Bansal M, Califano A, Estrada L, Schnell S, Irish JM, Massion PP, Quaranta V (2017) Novel hybrid phenotype revealed in small cell lung cancer by a transcription factor network model that can explain tumor heterogeneity. Cancer Res 77(5):1063–1074
Zeigler AC, Richardson WJ, Holmes JW, Saucerman JJ (2016) A computational model of cardiac fibroblast signaling predicts context-dependent drivers of myofibroblast differentiation. J Mol Cell Cardiol 94:72–81
Vasaikar SV, Ghosh S, Narain P, Basu A, Gomes J (2015) HSP70 mediates survival in apoptotic cells-Boolean network prediction and experimental validation. Front Cell Neurosci 9:319
Singh A, Nascimento JM, Kowar S, Busch H, Boerries M (2012) Boolean approach to signalling pathway modelling in HGF-induced keratinocyte migration. Bioinformatics 28(18):i495–i501
Verlingue L, Dugourd A, Stoll G, Barillot E, Calzone L, Londoño-Vallejo A (2016) A comprehensive approach to the molecular determinants of lifespan using a Boolean model of geroconversion. Aging Cell 15(6):1018–1026
Calzone L, Tournier L, Fourquet S, Thieffry D, Zhivotovsky B, Barillot E, Zinovyev A (2010) Mathematical modelling of cell-fate decision in response to death receptor engagement. PLoS Comput Biol 6(3):e1000702
Huang S, Eichler G, Bar-Yam Y, Ingber DE (2005) Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys Rev Lett 94(12):128701
Kauffman SA (1993) The origins of order: self-organization and selection in evolution. Oxford University Press, Oxford
Waddington CH (2014) The strategy of the genes, vol 20. Routledge
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30
Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath G, Wu G, Matthews L (2005) Reactome: a knowledgebase of biological pathways. Nucleic Acids Res 33(suppl 1):D428–D432
Chen H, Sharp BM (2004) Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinform 5(1):147
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4(9):R60
Freeman LC (1979) Centrality in social networks conceptual clarification. Soc Netw 1(3):215–239
Joy MP, Brock A, Ingber DE, Huang S (2005) High-betweenness proteins in the yeast protein interaction network. Biomed Res Int 2:96–103
Freeman LC (1977) A set of measures of centrality based on betweenness. Sociometry 35–41
Newman ME (2003) The structure and function of complex networks. SIAM Rev 45(2):167–256
Watts DJ, Strogatz SH (1998) Collective dynamics of ‘small-world’networks. Nature 393(6684):440–442
Terfve C, Cokelaer T, Henriques D, MacNamara A, Goncalves E, Morris MK, van Iersel M, Lauffenburger DA, Saez-Rodriguez J (2012) CellNOptR: a flexible toolkit to train protein signaling networks to data using multiple logic formalisms. BMC Syst Biol 6(1):133
Shmulevich I, Dougherty ER, Kim S, Zhang W (2002) Probabilistic Boolean networks: a rule-based uncertainty model for gene regulatory networks. Bioinformatics 18(2):261–274
Müssel C, Hopfensitz M, Kestler HA (2010) BoolNet—an R package for generation, reconstruction and analysis of Boolean networks. Bioinformatics 26(10):1378–1380
Trairatphisan P, Mizera A, Pang J, Tantar AA, Sauter T (2014) optPBN: an optimisation toolbox for probabilistic Boolean networks. PLoS ONE 9(7):e98001
Shmulevich I, Dougherty ER, Zhang W (2002) From Boolean to probabilistic Boolean networks as models of genetic regulatory networks. Proc IEEE 90(11):1778–1792
Brun M, Dougherty ER, Shmulevich I (2005) Steady-state probabilities for attractors in probabilistic Boolean networks. Signal Process 85(10):1993–2013
Ching WK, Zhang S, Ng MK, Akutsu T (2007) An approximation method for solving the steady-state probability distribution of probabilistic Boolean networks. Bioinformatics 23(12):1511–1518
Trairatphisan P, Mizera A, Pang J, Tantar AA, Schneider J, Sauter T (2013) Recent development and biomedical applications of probabilistic Boolean networks. Cell Commun Signal 11:46
Lommel MJ, Trairatphisan P, Gabler K, Laurini C, Muller A, Kaoma T, Vallar L, Sauter T, Schaffner-Reckinger E (2016) L-plastin Ser5 phosphorylation in breast cancer cells and in vitro is mediated by RSK downstream of the ERK/MAPK pathway. FASEB J 30(3):1218–1233
Klamt S, Saez-Rodriguez J, Gilles ED (2007) Structural and functional analysis of cellular networks with Cell NetAnalyzer. BMC Syst Biol 1(1):2
Naldi A, Berenguier D, Fauré A, Lopez F, Thieffry D, Chaouiya C (2009) Logical modelling of regulatory networks with GINsim 2.3. Biosystems 97(2):134–139
Krumsiek J, Pölsterl S, Wittmann DM, Theis FJ (2010) Odefy-from discrete to continuous models. BMC Bioinform 11(1):233
Di Cara A, Garg A, De Micheli G, Xenarios I, Mendoza L (2007) Dynamic simulation of regulatory networks using SQUAD. BMC Bioinform 8(1):462
Albert R, Thakar J (2014) Boolean modeling: a logic-based dynamic approach for understanding signaling and regulatory networks and for making useful predictions. Wiley Interdiscip Rev Syst Biol Med 6(5):353–369
Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30(1):207–210
Akutsu T, Kuhara S, Maruyama O, Miyano S (1998) A system for identifying genetic networks from gene expression patterns produced by gene disruptions and overexpressions. Genome Inform 9:151–160
Zhao Q (2005) A remark on” Scalar equations for synchronous Boolean networks with biological Applications” by C. Farrow, J. Heidel, J. Maloney, and J. Rogers. IEEE Trans Neural Netw 16(6):1715–1716
Bastolla U, Parisi G (1998) Relevant elements, magnetization and dynamical properties in Kauffman networks: a numerical study. Physica D 115(3):203–218
Bilke S, Sjunnesson F (2001) Stability of the Kauffman model. Phys Rev E 65(1):016129
Richardson KA (2005) Simplifying boolean networks. Adv Complex Syst 8(04):365–381
Dubrova E, Teslenko M (2011) A SAT-based algorithm for finding attractors in synchronous boolean networks. IEEE/ACM Trans Comput Biol Bioinf 8(5):1393–1399
Klamt S, Saez-Rodriguez J, Lindquist JA, Simeoni L, Gilles ED (2006) A methodology for the structural and functional analysis of signaling and regulatory networks. BMC Bioinform 7(1):56
Saadatpour A, Albert R, Reluga TC (2013) A reduction method for Boolean network models proven to conserve attractors. SIAM J Appl Dyn Syst 12(4):1997–2011
Zañudo JG, Albert R (2013) An effective network reduction approach to find the dynamical repertoire of discrete dynamic networks. Chaos: an Interdisciplinary. J Nonlinear Sci 23(2):025111
Veliz-Cuba A (2011) Reduction of Boolean network models. J Theor Biol 289:167–172
Naldi A, Remy E, Thieffry D, Chaouiya C (2011) Dynamically consistent reduction of logical regulatory graphs. Theoret Comput Sci 412(21):2207–2218
Huang S, Ingber DE (2000) Shape-dependent control of cell growth, differentiation, and apoptosis: switching between attractors in cell regulatory networks. Exp Cell Res 261(1):91–103
Schenone M, Dancik V, Wagner BK, Clemons PA (2013) Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol 9(4):232–240
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet J-P, Subramanian A, Ross KN (2006) The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313(5795):1929–1935
Woo JH, Shimoni Y, Yang WS, Subramaniam P, Iyer A, Nicoletti P, Martínez MR, López G, Mattioli M, Realubit R (2015) Elucidating compound mechanism of action by network perturbation analysis. Cell 162(2):441–451
Noh H, Shoemaker JE, Gunawan R (2017) Network perturbation analysis of gene transcriptional profiles reveals protein targets and mechanism of action of drugs and influenza A viral infection. bioRxiv:175364
Bansal M, Yang J, Karan C, Menden MP, Costello JC, Tang H, Xiao G, Li Y, Allen J, Zhong R (2014) A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol 32(12):1213–1222
Magkoufopoulou C, Claessen S, Tsamou M, Jennen D, Kleinjans J, van Delft J (2012) A transcriptomics-based in vitro assay for predicting chemical genotoxicity in vivo. Carcinogenesis 33(7):1421–1429
Kubicek S, Gilbert JC, Fomina-Yadlin D, Gitlin AD, Yuan Y, Wagner FF, Holson EB, Luo T, Lewis TA, Taylor B (2012) Chromatin-targeting small molecules cause class-specific transcriptional changes in pancreatic endocrine cells. Proc Natl Acad Sci 109(14):5364–5369
Thomas R (1991) Regulatory networks seen as asynchronous automata—a logical description. J Theor Biol 153(1):1–23
Harvey I, Bossomaier T Time out of joint: Attractors in asynchronous random boolean networks. In: Proceedings of the Fourth European Conference on Artificial Life, 1997. MIT Press, Cambridge, pp 67–75
Chaves M, Albert R, Sontag ED (2005) Robustness and fragility of Boolean models for genetic regulatory networks. J Theor Biol 235(3):431–449
Chaves M, Sontag ED, Albert R (2006) Methods of robustness analysis for Boolean models of gene control networks. Syst Biol (Stevenage) 153(4):154–167
Kraeutler MJ, Soltis AR, Saucerman JJ (2010) Modeling cardiac beta-adrenergic signaling with normalized-Hill differential equations: comparison with a biochemical model. BMC Syst Biol 4:157
Kirouac DC, Du J, Lahdenranta J, Onsum MD, Nielsen UB, Schoeberl B, McDonagh CF (2016) HER2+ cancer cell dependence on PI3K vs. MAPK signaling axes is determined by expression of EGFR, ERBB3 and CDKN1B. PLoS Comput Biol 12(4):e1004827
Nanavati C (2016) Pharmacodynamic systems analysis of HDAC and proteasome inhibition in multiple myeloma. State University of New York at Buffalo, Buffalo
Ramakrishnan V, Mager DE (2016) Abstracts accepted for American Conference on pharmacometrics 2016 (ACoP7): network-based analysis of pharmacodynamic heterogeneity in multiple myeloma cells. J Pharmacokinet Pharmacodyn 43(Suppl 1):11–122
Klemm K, Bornholdt S (2005) Stable and unstable attractors in Boolean networks. Phys Rev E 72(5 Pt 2):055101
Willadsen K, Wiles J (2007) Robustness and state-space structure of Boolean gene regulatory models. J Theor Biol 249(4):749–765
Fumia HF, Martins ML (2013) Boolean network model for cancer pathways: predicting carcinogenesis and targeted therapy outcomes. PLoS ONE 8(7):e69008
Irurzun-Arana I, Pastor JM, Trocóniz IF, Gómez-Mantilla JD (2017) Advanced Boolean modeling of biological networks applied to systems pharmacology. Bioinformatics (Oxford, England) 33 (7):1040
Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, Gonzalez-Baron M (2004) PI3K/Akt signalling pathway and cancer. Cancer Treat Rev 30(2):193–204
Klamt S, Gilles ED (2004) Minimal cut sets in biochemical reaction networks. Bioinformatics 20(2):226–234
Klamt S (2006) Generalized concept of minimal cut sets in biochemical networks. Biosystems 83(2–3):233–247
Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Richardson PG, Hideshima T, Munshi NC, Treon SP, Anderson KC (2002) Apoptotic signaling induced by immunomodulatory thalidomide analogs in human multiple myeloma cells: therapeutic implications. Blood 99(12):4525–4530
Richardson PG, Weller E, Lonial S, Jakubowiak AJ, Jagannath S, Raje NS, Avigan DE, Xie W, Ghobrial IM, Schlossman RL, Mazumder A, Munshi NC, Vesole DH, Joyce R, Kaufman JL, Doss D, Warren DL, Lunde LE, Kaster S, Delaney C, Hideshima T, Mitsiades CS, Knight R, Esseltine DL, Anderson KC (2010) Lenalidomide, bortezomib, and dexamethasone combination therapy in patients with newly diagnosed multiple myeloma. Blood 116(5):679–686
Zanudo JG, Albert R (2015) Cell fate reprogramming by control of intracellular network dynamics. PLoS Comput Biol 11(4):e1004193
Steinway SN, Zanudo JGT, Michel PJ, Feith DJ, Loughran TP, Albert R (2015) Combinatorial interventions inhibit TGFbeta-driven epithelial-to-mesenchymal transition and support hybrid cellular phenotypes. NPJ Syst Biol Appl 1:15014
Zhang R, Shah MV, Yang J, Nyland SB, Liu X, Yun JK, Albert R, Loughran TP Jr (2008) Network model of survival signaling in large granular lymphocyte leukemia. Proc Natl Acad Sci USA 105(42):16308–16313
Helikar T, Kowal B, McClenathan S, Bruckner M, Rowley T, Madrahimov A, Wicks B, Shrestha M, Limbu K, Rogers JA (2012) The cell collective: toward an open and collaborative approach to systems biology. BMC Syst Biol 6(1):96
Le Novere N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, Li L, Sauro H, Schilstra M, Shapiro B (2006) BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res 34(suppl 1):D689–D691
Klarner H, Streck A, Siebert H (2017) PyBoolNet: a python package for the generation, analysis and visualization of boolean networks. Bioinformatics 33(5):770–772
Naldi A, Monteiro PT, Müssel C, Kestler HA, Thieffry D, Xenarios I, Saez-Rodriguez J, Helikar T, Chaouiya C (2015) Cooperative development of logical modelling standards and tools with CoLoMoTo. Bioinformatics:btv013
Traynard P, Tobalina L, Eduati F, Calzone L, Saez-Rodriguez J (2017) Logic modeling in quantitative systems pharmacology. CPT Pharmacomet Syst Pharmacol 6(8):499–511
Knight-Schrijver VR, Chelliah V, Cucurull-Sanchez L, Le Novere N (2016) The promises of quantitative systems pharmacology modelling for drug development. Comput Struct Biotechnol J 14:363–370
Bloomingdale P, Housand C, Apgar JF, Millard BL, Mager DE, Burke JM, Shah DK (2017) Quantitative systems toxicology. Curr Opin Toxicol 4:79–87
Munos B (2009) Lessons from 60 years of pharmaceutical innovation. Nat Rev Drug Discov 8(12):959–968
Acknowledgements
This work was funded, in part, by NIH Grant GM57980.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table. S3
Summary of selected Boolean network models for various human physiological and pathophysiological systems. A brief description of network construction, reduction/refinement, validation, analyses, and findings are presented. Networks are highlighted based upon the physiological system that they represent: immune system (blue), breast (pink), gastrointestinal (brown), liver (green), lung (red), and uncategorized (gray). Supplementary material 4 (XLSX 59 kb)
Rights and permissions
About this article
Cite this article
Bloomingdale, P., Nguyen, V.A., Niu, J. et al. Boolean network modeling in systems pharmacology. J Pharmacokinet Pharmacodyn 45, 159–180 (2018). https://doi.org/10.1007/s10928-017-9567-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-017-9567-4