Abstract
This study aims to design epitope-based peptides for the utility of vaccine development by targeting Glycoprotein 2 (GP2) and Viral Protein 24 (VP24) of the Ebola virus (EBOV) that, respectively, facilitate attachment and fusion of EBOV with host cells. Using various databases and tools, immune parameters of conserved sequences from GP2 and VP24 proteins of different strains of EBOV were tested to predict probable epitopes. Binding analyses of the peptides with major histocompatibility complex (MHC) class I and class II molecules, population coverage, and linear B cell epitope prediction were peroformed. Predicted peptides interacted with multiple MHC alleles and illustrated maximal population coverage for both GP2 and VP24 proteins, respectively. The predicted class-I nonamers, FLYDRLAST, LFLRATTEL and NYNGLLSSI were found to cover the maximum number of MHC I alleles and showed interactions with binding energies of −7.8, −8.5 and −7.7 kcal/mol respectively. Highest scoring class II MHC binding peptides were EGAFFLYDRLASTVI and SPLWALRVILAAGIQ with binding energies of −6.2 and -5.6 kcal/mol. Putative B cell epitopes were also found on 4 conserved regions in GP2 and two conserved regions in VP24. Our in silico analysis suggests that the predicted epitopes could be a better choice as universal vaccine component against EBOV irrespective of different strains and should be subjected to in vitro and in vivo analyses for further research and development.
Similar content being viewed by others
Introduction
The recent outbreak of Ebola virus in Sub-Saharan Africa has once again proven that viral diseases pose a major threat to human society. There is no effective cure for most of the viral diseases. Most of them are treated symptomatically and death toll is always high. The only real action plan against viral diseases is the development of effective vaccines to prevent them. Vaccines are being constantly developed for viral diseases and many effective vaccines exist against yellow fever, measles, rubella, mumps, hepatitis B, influenza, human papillomavirus, polio, rabies etc. Still many old and newly emerging viral diseases remain a threat without proper vaccination. Examples include HIV (Human Immunodeficiency Virus) and Ebola virus.
Ebola virus belongs to the Group V (−)ssRNA, Order Mononegavirales, Family Filoviridae, Genus Ebolavirus, and Species Zaire ebolavirus. It was first identified in Democratic Republic of Congo (formerly Zaire) and it is named as such. It was first suspected to be a new strain of the closely related Marburg virus but was renamed to Ebola virus in 2010 (Feldmann et al. 2003; Peters et al. 1995; Ascenzi et al. 2008). The recent outbreak of the virus in West Africa has been responsible for more than 10,000 casualties so far (WHO 2015). Fruit bats are considered as the natural host of the virus and it is transmitted mainly through bodily fluids to human beings and other primates (Leroy et al. 2005; Pourrut et al. 2005; Funk and Kumar 2015; Drazen et al. 2014). Ebola virus disease (EVD), also known as Ebola hemorrhagic fever is a severe illness in humans. It is fatal without proper treatment. Recovery chances are really low, with the reported mortality rates being as high as 90 % (Sanchez et al. 2006). The current outbreak in West Africa has mortality rate of 70 % (WHO Ebola Response Team 2014). Ebola spreads through humans via direct contact with the blood, secretions, organs or other bodily fluids of infected people, and with objects contaminated with these fluids like bedding or clothing. According to a report published by the World Health Organisation (WHO) in September, 2014, health-care workers are frequently infected while treating patients with EVD. This occurs through close contact with patients without adequate precautions. Burial ceremonies can also play a role in the transmission of Ebola. Men who have recovered from the disease can still transmit the virus through their semen for up to 7 weeks after recovery from illness. Women can transmit the virus to children through breast milk. Symptoms of the disease occur in a specific order. First discernible symptoms are fever fatigue, muscle pain, sore throat and headache. This is followed by vomiting, diarrhea, rash, symptoms of impaired kidney and liver function, both internal and external bleeding. Laboratory findings include low white blood cell and platelet counts and elevated liver enzymes.
For prevention and cure, few vaccines have been developed and tested on non-human primates. These vaccines are either attenuated recombinant vesicular stomatitis virus vectors expressing the EBOV glycoprotein or an adenoviral vector encoding the Ebola glycoprotein (GP) (Sullivan et al. 2006; Geisbert et al. 2008; Jones et al. 2005). Both of these vaccines have been found promising in initial testing on non-human primates. These results demonstrate that it is indeed possible to develop a vaccine against Ebola virus (Sullivan et al. 2000, 2003). Currently, experimental drug treatments are being made available to impede Ebola outbreak. The major ones include; Zmapp, a mixture of three monoclonal antibodies that attack proteins on the surface of the virus (Qiu et al. 2014). Another drug TKM-Ebola has been designed to target strands of genetic material of the virus (Geisbert et al. 2010). The drug interrupts the genetic code of the virus and prevents it from making disease-causing proteins (Keller and Stiehm 2000). The US-based pharmaceutical company, Sarepta therapeutics, has developed a similar RNA treatment (Iversen et al. 2012). These drugs have tested on a small number of healthy volunteers but rarely on human patients. So far, no drug or vaccine has been approved by the FDA for the treatment.
Using existing knowledge about the structure and function of Ebola genome, Glycoprotein 2 (GP2) and Viral protein 24 (VP24) have been chosen as targets for vaccine development against this deadly virus (Lee et al. 2008; Huang et al. 2002). GP2 subunit of the virus has been found to be responsible for fusion of viral and host cell membrane (Volchkov et al. 1998). Cyrstallography studies have revealed that GP2 contains a central triple-stranded coiled coil followed by a disulfide-bonded loop which is homologous to an immunosuppressive sequence in retroviral glycoproteins (Malashkevich et al. 1999; Weissenhorn et al. 1998). The fusion peptides near the N termini form disulfide-bonded loops at one end of the molecule and that the C-terminal membrane anchors are at the same end, which possibly may help in initiation of fusion of membranes (Weissenhorn et al. 1998; Takada et al. 1997). The fusion active conformation of the subunit resembles to that of other viruses such as HIV and Influenza (Weissenhorn et al. 1998; Lee and Saphire 2009).
VP24 is a secondary matrix protein and is a minor component of virions. It possesses structural features commonly associated with viral matrix proteins (Han et al. 2003). It is chiefly responsible for the virus being able to evade the antiviral immune response of the body by suppressing the interferon (IFN) production. VP24 has been shown to compete with STAT1 to bind karyopherin α1, blocking nuclear accumulation and leading to inhibition of IFN signaling (Reid et al. 2006; Amarasinghe et al. 2014). VP24 is also responsible for correct assembly of a functional nucleocapsid and plays a role in virus assembly and budding (Han et al. 2003).
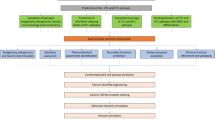
Biochemical, serological, and microbiological methods have been used to dissect pathogens and identify the components useful for vaccine development. Since the most abundant proteins are most often not suitable vaccine candidates, and the genetic tools required to identify the less abundant components maybe inadequate or not available at all, this approach can take years or even decades (Sette and Rappuoli 2010). In 1995, J. Craig Venter published the genome of the first free living organism, Haemophilus influenzae (a pathogenic bacterium) (Fleischmann et al. 1995). This opened a new way of using computers to rationally design vaccines by using the information present in the genome without going through the traditional microbiological and biochemical route. This new approach was called “Reverse Vaccinology” (Sette and Rappuoli 2010). The first example of reverse vaccinology approach was the development of a vaccine against serogroup B Neisseria meningitidis (MenB),a pathogen that causes 50 % of the meningococcal meningitis worldwide. In this study, bioinformatics methods were first used to screen the complete genome of MenB strain MC58, for genes encoding putative surface exposed or secreted proteins. In total, 350 novel vaccine candidates were predicted and expressed in E. coli; 28 were found to elicit protective immunity. It took less than 18 months to identify more and some novel vaccine candidates in MenB than had been discovered during the past 40 years by conventional methods (Pizza et al. 2000).
The approach of computers in this way for vaccine design is termed as “Immunoinformatics”. It mostly focuses on the design and study of algorithms for mapping potential B cell and T-cell epitopes, hence speeding up the time and lowering the cost needed for laboratory analysis of pathogen gene products (Doytchinova et al. 2003; Patronov and Doytchinova 2013). This concept also provides us with the concept of the “Immunome”, which can be defined as the set of antigens or epitopes that interface with the host immune system (Sette et al. 2005; De Groot and Berzofsky 2005). Thus immunomics bridges the discipline of genomics and proteomics by involving the immune system and focuses on elucidating the set of antigens that interact with the host immune system and the mechanisms involved in these interactions (Rinaudo et al. 2009).
Materials and Methods
Retrieval of Protein Sequences
The required protein sequences of GP2 and VP24 proteins from various strains of the Ebola virus. Table 1 lists all the sequences along with strains and Uniprot/GenBank accession numbers. The protein sequences belong to all the strains that have been found in various Ebola virus incidents throughout the world since its discovery. The information regarding different strains and their proteomes was acquired from Viral Bioinformatics Resource Center (www.biovrus.org). The sequences were stored as two fasta files containing multiple sequences for each protein respectively.
Control
The matrix protein 1 [Influenza A virus (H5N1)] was taken as control as it a well-studied viral antigen showing proper immune response in humans. It has been tested as an adjuvanted virosomal H5N1 vaccine and found to induce a balanced Th1/Th2 CD4(+) T cell response in man (Pederson et al. 2014). It was subjected to all the in silico procedures in this study to prove that the pipeline is adequate for antigenic predictions.
Multiple Sequence Alignment (MSA)
Multiple sequence alignment was used to identify conserved regions in the protein sequences of GP2 and VP24 respectively. Amino acid sequences which are found to be conserved among different strains signify less ability of the protein to mutate in that region. They also provide good starting point for the analysis of antigenic sequences and lymphocytic epitopes since any vaccine designed using such conserved sequences should work on all the strains. PRALINEWWW was used to perform multiple alignment of the proteins (Simossis and Heringa 2005). The tool is available in form of a web application at “www.ibi.vu.nl/programs/pralinewww/”. Praline is a highly customizable MSA application (Simossis and Heringa 2005; Heringa 2002). It provides many different alignment strategies such as progressive alignment, integration of structural features such as secondary structures and trans-membrane regions (Simossis and Heringa 2005). The output can be obtained in tree form or in the form of a fasta file containing the alignment. For our purpose, we used BLOSUM62 as the weight matrix for the alignment. Gap opening and extension penalties were chosen to be 12 and 1 respectively. The alignment strategy incorporated was PSI-BLAST pre-profile processing (Homology-extended alignment) with 3 PSI-BLAST iterations at an E-value cut-off of 0.01 (Simossis et al. 2005). The alignment was made against the NCBI NR (non-redundant) database. DSSP-defined secondary structure searching along with secondary structure prediction using PSIPRED was also used (Heringa 1999). And the output was generated as a fasta file containing the multiple alignment.
Antigenicity Prediction
Antigenicity prediction of all the conserved sequences generated in the previous step was performed to assess their overall possible role in generating immune response. Vaxijen server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was used as it does not rely on sequence similarities with known antigens (Doytchinova and Flower 2007). This provides with a unique insight into potentially novel antigenic sequences which may not have obvious sequence similarities. This also makes it a very useful tool for small sequences (as in this case), since sequence similarity predictions depend on the overall length of the sequences. And small sequences may generate many localized hits which may be irrelevant. The vaxijen server gives results in form of probability scores, prediction threshold for which was kept at 0.5 for getting the accuracy of 87 % (Gededzha et al. 2014).
T-Cell Epitope Prediction for MHC I and MHC II
The involvement of short sequences of amino acids in many processes of molecular biology such as the binding of immunogenic peptides to major histocompatibility complex (MHC) molecules is well established. Reliable predictions of immunogenic peptides can minimize the experimental effort needed to identify new epitopes to be used in vaccine design. NetCTL (http://www.cbs.dtu.dk/services/NetCTL/) is a web-based tool for predicting human cytotoxic T lymphocyte (CTL) epitopes in any given protein. It does so by integrating predictions of proteasomal cleavage, TAP transport efficiency, and MHC class I affinity. It is a highly sensitive tool which performs better than other tools in large scale comparisons (Larsen et al. 2007). It includes 12 MHC I supertypes in its prediction protocol. 0.15 was used as the threshold for C terminal cleavage, 0.05 for TAP transport efficiency and 0.5 for epitope prediction as these values increase sensitivity to a larger extent than they lower the specificity of the prediction (Nielsen et al. 2003, 2005; Peters et al. 2003). The peptides which were selected in the anitgenicity prediction were used as the input.
Antigen presenting cells (APCs) present peptides from the extra cellular space to T helper cells, which are activated if the peptides are recognized as non-self. The peptides are presented on the cell surface in complex with major histocompatibility class II (MHC II) molecules. Thus, prediction of peptides that bind to MHC II is a very important step for in silico vaccine design. The MHC class II binding groove, being open at both ends, makes the correct alignment of a peptide in the binding groove an important part of identifying the core of an MHC class II binding peptide. MHC II binding prediction was performed by using the IEDB MHC II prediction tool at http://tools.immuneepitope.org/mhcii/. It uses a novel Stabilization matrix alignment method (SMM-align) (Nielsen et al. 2007).
B-Cell Epitope Prediction
One of the key aspects of the immune system is the antibody-mediated identification of foreign, infectious objects, such as bacteria and viruses. Antibodies bind to antigens at sites known as B-cell epitopes. Ability to identify these binding areas in the antigen sequence or on its surface is important for the development of vaccines. The linear B-cell epitope is a short segment in the amino acid sequence of the antigen. There are structural B cell epitiopes which are non-contiguous and depend on the 3D structure of the antigens. The reason for linear epitope prediction is that it is computationally more feasible. Since this work is based on sequence information and does not focus on the structures of GP2 and VP24, we focus on the conserved linear peptides of the proteins which are established through MSA and not on the structural information. LBTope (http://www.imtech.res.in/raghava/lbtope/index.php) was used to perform this prediction. It uses very large datasets of 14,876 B-cell epitope and 23,321 non-epitopes of variable length, 12,063 B-cell epitopes and 20,589 non-epitopes of fixed length and 1042 epitopes and 1795 non-epitopes where each epitope or non-epitope has been experimentally validated (Singh et al. 2013). Availability of large datasets increases the validity of machine-learning predictive algorithms such as ANN. To increase the specificity of prediction, the probability was increased to 80 %.
Population Coverage Analysis of MHC I Epitopes
The MHC is a highly polymorphic group of genes. Different populations typically express different repertoires of MHC alleles. An epitope-based vaccine can include only a limited number of peptides due to economic and regulatory issues. Hence, it is very important to identify the optimal set of peptides for a vaccine. Constraints such as peptide mutation rates and maximum number of selected peptides place an additional burden on the overall design process (Toussaint and Kohlbacher 2009). Optitope (http://etk.informatik.uni-tuebingen.de/optitope) predicts the overall immunogenicity of a peptide set which depends on the individual immunogenicities of each peptide with respect to the MHC alleles in a given population. North Africa, South East Asia, South West Asia, Sub Saharan Africa were reported by the WHO to have Ebola virus breakout sometimes in the past. These populations were used for the analysis. The analysis focused on MHC I because of the fact that viral peptides are presented only on MHC I via the endogenous pathway.
Docking of the Selected Epitopes with MHC Alleles
Molecular docking is a key tool in structural molecular biology and computer-assisted drug design. The goal of ligand–protein docking is to predict the predominant binding mode of a ligand with a protein of known three-dimensional structure. It is a hypothesis generating procedure which provides a basis for further in vitro/in vivo analysis. It was used to analyze the binding of the selected MHC I and MHC II epitopes with the 3D structures of respective MHC molecules. The epitopes were selected for both MHC I and MHC II on the basis of their binding scores in the predictions. These epitopes were modeled by using the short peptide folding method of PEP-FOLD at the mobyle web server (Thévenet et al. 2012; Maupetit et al. 2009, 2010). The 3D structures of the MHC I HLA-A2 (pdb id: 3MRE) and MHC2 HLA-DR1(pdb id: 1AQD) were downloaded from the PDB server and modified for further usage as receptors.
Autodock vina, developed by the Scripps Research Institute, was used for docking the epitopes with the MHC molecules (Trott and Olson 2010). It was developed as an improvement over the original autodock program both in term of accuracy and speed. Autodock vina can use multicore processors and hence is much faster than the original autodock.
Results
Selection of Conserved Sequences
Multiple sequence alignment by PRALINEWWW led to the discovery of conserved sequences in the GP2 and VP24 proteins. 13 conserved regions were found in GP2. Out of these, two were discarded for being too short (nine and seven residues respectively). The selection criterion was the conservation rating of seven or above in the PRALINEWWW conservation colour chart. Secondary structure info generated by PRALINEWWW (PSIPRED) was checked to ensure that there were no gaps in the defined helices or strands. Secondary structure info was annotated in the fasta headers of the conserved sequences.
11 conserved regions were found in VP24. Out of these, one was discarded for being too short (seven residues). The selection criterion was kept the same. In this case most of the sequences were edited to avoid introducing gaps in conserved helices and strands according to the secondary structure info generated. Table 2 lists the conserved sequences found in both the proteins.
Antigenicity of the Conserved Sequences
Analysis revealed that eight and five conserved sequences, respectively, from GP2 and VP24 proteins met the criteria of default threshold level, ≥0.5, in VaxiJen as listed in Table 2. The control antigen also tested positive in the vaxijen server.
T-Cell Epitope Prediction for MHC I and MHC II
NetCTL prediction tool covering all supertypes created a total of 160 and 34 nonamers from the conserved sequences of GP2 and VP24 proteins, respectively based on the tool’s combined score threshold. Further analysis revealed 76 unique epitopes reacting with 12 MHC I alleles in GP2 and 19 unique epitopes reacting with 12 MHC I alleles in VP24 as listed in Table 3. In the control antigen, 188 MHC I and 123 MHC II epitopes were predicted using the same tools.
IEDB MHC II epitope prediction tool generated 72 unique binding peptides from the GP2 protein having affinity values <250 nM which reacted to 36 unique HLA DP, DQ, and DR alleles and 40 unique binding peptides from the VP24 protein having affinity values <250 nM which reacted to 30 unique HLA DP, DQ, and DR alleles as listed in Table 4.
B-Cell Epitope Prediction
According to the criteria set for the prediction of B cell epitopes using LBTope server along with the basis of VaxiJen scores, GP2 protein was predicted to have four conserved peptides and VP24 protein was predicted to have two conserved peptides to contain B-cell epitopes. Similar criteria set was used for the control antigen and 31 B cell epitopes were predicted by LBTope.
Population Coverage Analysis of MHC I Epitopes
Over a thousand different human MHC (HLA) alleles are known and different HLA types are expressed at different frequencies in different ethnicities. Identified epitopes that bind to several MHC alleles would be considered as the best probable epitope only if their combined frequency in a population shows good coverage by approaching 100 % or close to 100 %.
Opitope prediction server found FLYDRLAST, LFLRATTEL from the GP2 protein and NYNGLLSSI from the VP24 protein. FLYDRLAST showed interaction with HLA-A*0201 in North African, South West Asian and Sub Saharan African populations and LFLRATTEL showed interaction with HLA-A*2402 in South East Asian Population. NYNGLLSSI showed interaction with both HLA-A*0201 and HLA-A*2402 in all the four target populations.
Docking of the Selected Epitopes with MHC Alleles
Using AutoDock Vina, binding models of predicted epitopes to their respective HLA molecules (both class I and class II) were generated (Figs. 1, 2). In case of class 1, epitopes FLYDRLAST and LFLRATTEL from GP2 protein bound to the binding groove of HLA-A2 (pdb id 3MRE) with the binding energies of −7.8 and −8.5 kcal/mol respectively (Fig. 1a, b). Furthermore, FLYDRLAST forms a single hydrogen bond with the residue ASP77 having a bond length of 2.07 Angstrom and LFLRATTEL forms three hydrogen bonds with residues ASP77, LYS146 and ARG97 having bond lengths of 1.812, 2.143 and 2.145 Angstrom respectively. The epitope NYNGLLSSI from the VP24 protein bound to the binding groove with the binding energy of −7.7 kcal/mol (Fig. 1c) and forms two hydrogen bonds with residues ASP77 and HIS114 having bond lengths of 2.108 and 2.249 Angstrom respectively. The selected MHC I epitope from the control antigen, GMLGFVFTL bound to the binding groove of the same HLA molecule as above. It showed a binding energy of −7.4 kcal/mol and also formed a single hydrogen bind with THR73 with a bond length of 2.182 Angstrom (Fig. 3a).
In case of class 2, epitope EGAFFLYDRLASTVI from GP2 protein bound to the binding groove of HLA-DR1 (pdb id 1AQD) with the binding energy of −6.2 kcal/mol (Fig. 2a). It forms four hydrogen bonds with residues ASN62, SER53, GLN9 and THR77 having bond lengths of 2.125, 2.165, 1.93 and 2.236 Angstrom respectively. The epitope SPLWALRVILAAGIQ from VP24 protein bound with the binding energy of −5.6 kcal/mol (Fig. 2b). It forms a single hydrogen bond with the residue ASN82 having a bond length of 2.056 Angstrom. The selected MHC II epitope from the control antigen, GLIYNRMGTVTTEVA bound to the binding groove with the binding energy of −5.2 kcal/mol and formed four hydrogen bonds with GLU55, SER53, ASN32 and GLN9. The lengths of the bonds are 1.987, 1.926, 1.967 and 2.157 Angstrom respectively (Fig. 3b).
Discussion
This study was focused on the in silico prediction of peptides from two proteins from the Ebola virus in the light of the recent outbreak. Envelope Glycoprotein (GP2) and Viral protein 24 (VP24) were selected as they have been shown to play immensely important role in the viral infection and evasion of the host immune system. Since the virus has mutated multiple times since the first known outbreak, 29 different sequences of GP2 and 19 different sequences of VP24 were downloaded. After performing multiple alignments of all the sequences each protein, 11 and 10 conserved regions were selected for further analysis. Eight and five sequences respectively from GP2 and VP24 were predicted as potential antigens by the VaxiJen server. These antigenic sequences formed the basis of all further analysis.
The major histocompatibilty complex (MHC) is a highly polymorphic group of cell-surface receptors found on all the cells of the body. MHC is categorized in two types: MHC I and II. MHC I is found on all nucleated cells of the body while MHC II is expressed only by APCs. Their role is to present short peptide sequences both from self-proteins and foreign proteins to other elements of the immune system (T Cells) and hence play perhaps the most important role in the training of the immune system. The peptides which are expressed on MHCs are called T Cell epitopes. The antigenic sequences selected previously were used to predict MHC I and II binding epitopes. GP2 yielded 76 unique epitopes which exhibited binding to 12 unique MHC I alleles and 72 unique epitopes for 36 unique MHC II alleles. 19 unique epitopes were found from VP24 for 12 unique MHC I alleles and 40 unique epitopes were found for 30 unique MHC II alleles.
MHC I epitopes predicted above were subjected to population coverage analysis as not all the MHC alleles are expressed in every population. This is necessary to select proper peptides for rational vaccine design since the peptides reacting with the most expressed MHC alleles in a target population will be the most appropriate. The target populations (North Africa, South West Asia, South East Asia and Sub Saharan Africa) selected for analysis were based on all of the outbreaks of Ebola virus since its discovery. The analysis revealed that FLYDRLAST epitope from GP2 reacted to HLA-A*0201, which is the most expressed human MHC I allele in North African, South West Asian and Sub Saharan African populations and LFLRATTEL epitope from the same protein reacted with HLA-A*2402, the most expressed allele in the South East Asian Population. From VP24, NYNGLLSSI epitope reacted with both HLA-A*0201 and HLA-A*2402 in all the target populations.
In addition to the TH Cell response mediated by MHC II, B Cells also play a role in humoral immunity. The peptide regions which bind to B cell receptors are called B cell epitopes. All the conserved sequences were used to perform B cell epitope prediction. Four conserved peptides from GP2 and 2 from VP24 were found to contain B cell epitopes.
The interaction of GP2 and VP24 with the human immune system is already established in various studies. The principle interaction in form of MHC I and II presentation was studied by using molecular docking to analyze the structural binding between MHC alleles and predicted peptides. Autodock vina showed that FLYDRLAST and LFLRATTEL from GP2 fit into the binding groove of with HLA-A2 (MHC I) with binding energies of −7.8 and −8.5 kcal/mol while NYNGLLSSI from VP24 fit into the binding groove of the same MHC molecule with binding energy of −5.6 kcal/mol. The MHC II binding groove is open from both ends unlike the MHC I binding groove which is closed. Therefore, longer peptides (up to about 15 residues long) can fit into this groove. Docking with vina showed that the peptide EGAFFLYDRLASTVI from GP2 fit into the binding groove of HLA-DR1 (MHC II) with the binding energy of −6.2 kcal/mol and SPLWALRVILAAGIQ from VP24 fit into the binding groove of the same molecule with the binding energy of −5.6 kcal/mol. These energies along with the formation of hydrogen bonds show that the selected epitopes can be used as a basis for a novel peptide based vaccine against most of the known strains of Ebola virus. And possibly against newly emerging strains too because the basis of this study was the conservation of protein sequences in various strains.
In this study, we have concentrated on the predicted peptides which can be used as vaccine candidates. Further studies will concentrate on the delivery mechanisms including various adjuvants (Freund’s adjuvant, liposomes, virosomes etc.) and the simulation of interactions with the immune system as a complete vaccine system. Development of polytopic vaccines is a also a valid strategy which takes advantage of linear peptide sequences.
Conclusion
The world is now the habitat of more than seven billion people. With the advent of medical technology, new kinds of diseases are also emerging along with new viruses. Developing world, in particular, is more affected by these sorts of diseases. Diseases which have earlier been recognized as zoonotic are now spreading from human to human. However, medical science has always tried to cope with the problems with the pace of replicating disease. New technologies involving high performance computing for “in silico” design of drugs and vaccines cut the time required to do so by a large extent. Bioinformatics has played an immensely important role in the diverse areas of life sciences. From the basic study of macromolecule sequences and structures to the application of machine-learning algorithms to simulate complete biological systems, bioinformatics is the central pillar of modern life science research. Almost all of the bioinformatics software is either available for free to use or the algorithms are in public domain and can be implemented in any programming language. This study focused on two things. Firstly the importance of bioinformatics and free software in the field of life science research but secondly and most importantly, it focused on the fact that there is a great need to apply bioinformatics to world-wide problems like the recent Ebola virus outbreak to provide with lifesaving solutions (diagnostics, drugs and vaccines) in a short amount of time to maximum people at the lowest possible costs.
References
Amarasinghe GK, Xu W, Edwards MR, Borek DM, Mittal A (2014) Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe 16(2):187–200. doi:10.1016/j.chom.2014.07.008
Ascenzi P, Bocedi A, Heptonstall J, Capobianchi MR, Di Caro A, Mastrangelo E, Bolognesi M, Ippolito G (2008) Ebolavirus and Marburgvirus: insight the Filoviridae family. Mol Asp Med 29(3):151–185. doi:10.1016/j.mam.2007.09.005
De Groot AS, Berzofsky JA (2005) From genome to vaccine—new immnoinformatics tools for vaccine design. Methods 34(4):425–428. doi:10.1016/j.ymeth.2004.06.004
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform 8(1):4. doi:10.1186/1471-2105-8-4
Doytchinova IA, Taylor P, Flower DR (2003) Proteomics in vaccinology and immunobiology: an informatics perspective of the immunone. J Biomed Biotechnol 5:267–290. doi:10.1155/s1110724303209232
Drazen JM, Kanapathipillai R, Campion EW, Rubin EJ, Hammer SM, Morrissey S, Baden LR (2014) Ebola and quarantine. N Engl J Med 371(21):2029–2030. doi:10.1056/NEJMe1413139
Feldmann H, Jones S, Klenk HD, Schnittler HJ (2003) Ebola virus: from discovery to vaccine. Nat Rev Immunol 3(8):677–685. doi:10.1038/nri1154
Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM et al (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269(5223):496–512. doi:10.1126/science.7542800
Funk DJ, Kumar A (2015) Ebola virus disease: an update for anesthesiologists and intensivists. Can J Anaesth 62(1):80–91. doi:10.1007/s12630-014-0257-z
Gededzha MP, Mphahlele MJ, Selabe SG (2014) Prediction of T-cell epitopes of hepatitis C virus genotype 5a. Virol J 11(1):187. doi:10.1186/1743-422x-11-187
Geisbert TW, Daddario-DiCaprio KM, Lewis MG, Geisbert JB, Grolla A et al (2008) Vesicular stomatitis virus-based Ebola vaccine is well-tolerated and protects immunocompromised nonhuman primates. PLoS Pathog 4(11):e1000225. doi:10.1371/journal.ppat.1000225
Geisbert TW et al (2010) Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet 375(9729):1896–1905. doi:10.1016/S0140-6736(10)60357-1
Han Z, Boshra H, Sunyer JO, Zwiers SH, Paragas J, Harty RN (2003) Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J Virol 77(3):1793–1800. doi:10.1128/JVI.77.3.1793-1800.2003
Heringa J (1999) Two strategies for sequence comparison: profile- preprocessed and secondary structure-induced multiple alignment. Comput Chem 23:341–364. doi:10.1016/s0097-8485(99)00012-1
Heringa J (2002) Local weighting schemes for protein multiple sequence alignment. Comput Chem 26:459–477. doi:10.1016/S0097-8485(02)00008-6
Huang Y, Xu L, Sun Y, Nabel GJ (2002) The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol Cell 10:307–316. doi:10.1016/S1097-2765(02)00588-9
Iversen PL, Warren TK, Wells JB et al (2012) Discovery and early development of AVI-7537 and AVI-7288 for the treatment of Ebola virus and Marburg virus infections. Viruses 4(11):2806–2830. doi:10.3390/v4112806
Jones SM, Feldmann H, Ströher U, Geisbert JB, Fernando L, Grolla A, Klenk H, Sullivan NJ, Volchkov VE, Fritz EA, Daddario KM, Hensley LE, Jahrling PB, Geisbert TW (2005) Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat Med 11:786–790. doi:10.1038/nm1258
Keller MA, Stiehm ER (2000) Passive immunity in prevention and treatment of infectious diseases. Clin Microbiol Rev 13(4):602–614. doi:10.1128/CMR.13.4.602-614.2000
Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M (2007) Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics 8(1):424. doi:10.1186/1471-2105-8-424
Lee JE, Saphire EO (2009) Ebolavirus glycoprotein structure and mechanism of entry. Futur Virol 4(6):621–635. doi:10.2217/fvl.09.56
Lee JE, Fusco ML, Oswald WB, Hessell AJ, Burton DR, Saphire EO (2008) Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182. doi:10.1038/nature07082
Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez J, Swanepoel R (2005) Fruit bats as reservoirs of Ebola virus. Nature 438:575–576. doi:10.1038/438575a
Malashkevich VN, Schneider BJ, Mcnally ML, Milhollen MA, Pang JX, Kim PS (1999) Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc Natl Acad Sci 96:2662–2667. doi:10.1073/pnas.96.6.2662
Maupetit J, Derreumaux P, Tufféry P (2009) PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res 37:W498–W508. doi:10.1093/nar/gkp323
Maupetit J, Derreumaux P, Tuffery P (2010) A fast and accurate method for large-scale de novo peptide structure prediction. J Comput Chem 31(4):726–738. doi:10.1002/jcc.21365
Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, Brunak S, Lund O (2003) Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12:1007–1017. doi:10.1110/ps.0239403
Nielsen M, Lundegaard C, Brunak S, Lund O, Kesmir C (2005) The role of the proteasome in generating cytotoxic T cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Imunogenetics 57(1–2):33–41. doi:10.1007/s00251-005-0781-7
Nielsen M, Lundegaard C, Lund O (2007) Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinform 8(1):238. doi:10.1186/1471-2105-8-238
Patronov A, Doytchinova I (2013) T-cell epitope vaccine design by immunoinformatics. Open Biol 3(1):120–139. doi:10.1098/rsob.120139
Pederson GK, Sjursen H, Nostbakken JK, Jul-Larsen A, Hoschler K, Cox RJ (2014) Matrix M(TM) adjuvanted virosomal H5N1 vaccine induces balanced Th1/Th2 CD4(+) T cell responses in man. Hum Vaccines Immunother 10(8):2408–2416. doi:10.4161/hv.29583
Peters CJ, Sanchez A, Rollin PE, Ksiazek TG, Murphy FA (1995) Filoviridae: Marburg and Ebola viruses. In: Fields BN (ed) Fields virology. Lippincott-Raven, Philadelphia, pp 1161–1176
Peters B, Bulik S, Tampe R, Endert PMV, Holzhutter HG (2003) Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol 171(4):1741–1749. doi:10.4049/jimmunol.171.4.1741
Pizza M, Scarlato V, Masignani V et al (2000) Identification of vaccine candidates against serogroup B meningococcus by whole genome sequencing. Science 287(5459):1816–1820. doi:10.1126/science.287.5459.1816
Pourrut X, Kumulungui B, Wittmann T, Moussavou G, Délicat A, Yaba P, Nkoghe D, Gonzalez JP, Leroy EM (2005) The natural history of Ebola virus in Africa. Microbes Infect 7(7–8):1005–1014. doi:10.1016/j.micinf.2005.04.006
Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB et al (2014) Reversion of advanced Ebola virus disease in nonhuman primates with Zmapp. Nature 514:47–53. doi:10.1038/nature13777
Reid S, Leung LW, Hartman AL, Martinez O, Shaw ML, Carbonnelle C, Volchkov VE, Nichol ST, Basler CF (2006) Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J Virol 80(11):5156–5167. doi:10.1128/JVI.02349-05
Rinaudo CD, Telford JL, Rappuoli R, Seib KL (2009) Vaccinology in the genomics era. J Clin Investig 119(9):2515–2525. doi:10.1172/JCI38330
Sanchez A, Geisbert TW, Feldmann H (2006) Filoviridae: Marburg and Ebola Viruses. In: Fields BN (ed) Fields virology. Lippincott Williams & Wilkins, Philadelphia, pp 1409–1448
Sette A, Rappuoli R (2010) Reverse vaccinology: developing vaccines in the era of genomics. Immunity 33(4):530–541. doi:10.1016/j.immuni.2010.09.017
Sette A, Fleri W, Peters B et al (2005) A roadmap for immunomics of category A-C pathogens. Immunity 22(2):155–161. doi:10.1016/j.immuni.2005.01.009
Simossis VA, Heringa J (2005) PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res 33:W289–W294. doi:10.1093/nar/gki390
Simossis VA, Kleinjung J, Heringa J (2005) Homology-extended sequence alignment. Nucleic Acids Res 33(3):816–824. doi:10.1093/nar/gki233
Singh H, Ansari HR, Raghava GPS (2013) Improved method for linear B-Cell epitope prediction using antigen’s primary sequence. PLoS One 8(5):e62216. doi:10.1371/journal.pone.0062216
Sullivan NJ, Sanchez A, Rollin PE, Yang Z, Nabel GJ (2000) Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605–609. doi:10.1038/35046108
Sullivan NJ, Geisbert TW, Geisbert JB, Xu L, Yang Z, Roederer M, Kou RA, Jahrling PB, Nabel GJ (2003) Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature 424:681–684. doi:10.1038/nature01876
Sullivan NJ, Geisbert TW, Geisbert JB, Shedlock DJ, Xu L, Lamoreaux L, Custers JHHV, Popernack PM, Yang Z, Pau MG, Roederer M, Koup RA, Goudsmit J, Jahrling PB, Nabel GJ (2006) Immune protection of nonhuman primates against Ebola virus with single low-dose adenovirus vectors encoding modified Gps. PLoS Med 3(6):865–873. doi:10.1371/journal.pmed.0030177
Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y (1997) A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci 94:14764–14769. doi:10.1073/pnas.94.26.14764
Thévenet P, Shen Y, Maupetit J, Guyon F, Derreumaux P, Tufféry P (2012) PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res 40(W1):W288–W293. doi:10.1093/nar/gks419
Toussaint NC, Kohlbacher O (2009) OptiTope—a web server for the selection of an optimal set of peptides for epitope-based vaccines. Nucleic Acids Research 37:W617–W622. doi:10.1093/nar/gkp293
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31(2):455–461. doi:10.1002/jcc.21334
Volchkov VE, Feldmann H, Volchkova VA, Klenk H (1998) Microbiology Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci 95:5762–5767. doi:10.1128/JVI.02486-06
Weissenhorn W et al (1998) Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell 2(5):605–616. doi:10.1016/S1097-2765(00)80159-8
WHO Ebola Response Team (2014) Ebola Virus Disease in West Africa—The first 9 months of the epidemic and forward projections. N Engl J Med 371:1481–1495. doi:10.1056/NEJMoa1411100
WHO-Ebola data and statistics—situation summary. http://apps.who.int/ebola/en/ebola-situation-reports. Accessed 9 April 2015
Acknowledgments
The authors sincerely thank Dr. Alok Lehri, Director, Central Instrumentation Facility, NBRI, Lucknow, India for his valuable guidance and comments during preparation of the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Authors declare that they have no conflict of interest.
Funding
This study was conducted using the resources available in the Bioinformatics Laboratory of Institute of Engineering and Technology, Sitapur Road, Lucknow and no additional funding was involved.
Ethical approval
The authors declare that there were no animals or humans involved in this study as subjects.
Rights and permissions
About this article
Cite this article
Srivastava, P.N., Jain, R., Dubey, S.D. et al. Prediction of Epitope-Based Peptides for Vaccine Development from Coat Proteins GP2 and VP24 of Ebola Virus Using Immunoinformatics. Int J Pept Res Ther 22, 119–133 (2016). https://doi.org/10.1007/s10989-015-9492-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-015-9492-6