Abstract
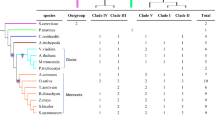
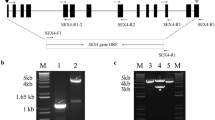
Starch branching enzyme (SBE) is one of the key enzymes involved in starch biosynthetic metabolism. In this study, six SBE family genes were identified from the cassava genome. Phylogenetic analysis divided the MeSBE family genes into dicot family A, B, C, and the new group. Tissue-specific analysis showed that MeSBE2.2 was strongly expressed in leaves, stems cortex, and root stele, and MeSBE3 had high expression levels in stem cortex and root stele of plants in the rapid growth stage under field condition, whereas the expression levels of MeSBE2.1, MeSBE4, and MeSBE5 were low except for in stems cortex. The transcriptional activity of MeSBE2.2 and MeSBE3 was higher compared with other members and gradually increased in the storage roots during root growth process, while the other MeSBE members normally remained low expression levels. Expression of MeSBE2.2 could be induced by salt, drought, exogenous abscisic acid, jasmonic acid, and salicylic acid signals, while MeSBE3 had positive response to drought, salt, exogenous abscisic acid, and salicylic acid in leaves but not in storage root, indicating that they might be more important in starch biosynthesis pathway under diverse environments.





Similar content being viewed by others
References
Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buleon A, Colonna P, Preiss J (1996) From glycogen to amylopectin a model for the biogenesis of plant starch granule. Cell 86:349–352
Smith AM, Denyer K, Martin C (1997) The synthesis of the starch granule. Ann Rev Plant Physiol Plant Mol Biol 48:67–87
Bhattacharyya MK, Smith AM, Ellis TH, Hedley C, Martin C (1990) The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell 60:115–122
Carciofi M, Blennow A, Jensen SL, Shaik SS, Henriksen A, Buleon A, Holm PB, Hebelstrup KH (2012) Concerted suppression of all starch branching enzyme genes in barley produces amylose-only starch granules. BMC Plant Biol 12:223. doi:10.1186/1471-2229-12-223
Haworth WN, Peat S, Bourne EJ (1944) Synthesis of amylopectin. Nature 154:236–238
Fisher DK, Gao M, Kim KN, Boyer CD, Guiltinan MJ (1996) Two closely related cDNAs encoding starch branching enzyme from Arabidopsis thaliana. Plant Mol Biol 30:97–108
Larsson CT, Khoshnoodi J, Ek B, Rask L, Larsson H (1998) Molecular cloning and characterization of starch-branching enzyme II from potato. Plant Mol Biol 37:505–511
Koda A, Bogaki T, Minetoki T, Hirotsune M (2005) High expression of a synthetic gene encoding potato alpha-glucan phosphorylase in Aspergillus niger. J Biosci Bioeng 100:531–537. doi:10.1263/jbb.100.531
Larsson C-T, Hofvander P, Khoshnoodi J, Ek B, Rask L, Larsson H (1996) Three isoforms of starch synthase and two isoforms of branching enzymes are present in potato tuber starch. Plant Sci 117:9–16
Sun C, Sathish P, Ek B, Deiber A, Jansson C (1996) Demonstration of in vitro starch branching enzyme activity for a 51/50-kDa polypeptide isolated from developing barley (Hordeum vulgare) caryopses. Physiol Plant 96:473–483
Gao M, Fisher DK, Kim KN, Shannon JC, Guiltinan MJ (1996) Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggests isoform specialization. Plant Mol Biol 30:1223–1232
Morell MK, Blennow A, Kosar-Hashemi B, Samuel MS (1997) Differential expression and properties of starch branching enzyme isoforms in developing wheat endosperm. Plant Physiol 113:201–208
Hawker JS, Ozbun JL, Ozaki H, Greenberg E, Preiss J (1974) Interaction of spinach leaf adenosine diphosphate glucose alpha-1,4-glucan alpha-4-glucosyl transferase and alpha-1,4-glucan, alpha-1,4-glucan-6-glycosyl transferase in synthesis of branched alpha-glucan. Arch Biochem Biophys 160:530–551
Burton RA, Bewley JD, Smith AM, Bhattacharyya MK, Tatge H, Ring S, Bull V, Hamilton WD, Martin C (1995) Starch branching enzymes belonging to distinct enzyme families are differentially expressed during pea embryo development. Plant J 7:3–15
Baguma Y, Sun C, Ahlandsberg S, Mutisya J, Palmqvist S, Rubaihayo PR, Magambo MJ, Egwang TG, Larsson H, Jansson C (2003) Expression patterns of the gene encoding starch branching enzyme II in the storage roots of cassava (Manihot esculenta Crantz). Plant Sci 164:833–839. doi:10.1016/s0168-9452(03)00072-4
Salehuzzaman SN, Jacobsen E, Visser RG (1992) Cloning, partial sequencing and expression of a cDNA coding for branching enzyme in cassava. Plant Mol Biol 20:809–819
Guan H, Li P, Imparl-Radosevich J, Preiss J, Keeling P (1997) Comparing the properties of Escherichia coli branching enzyme and maize branching enzyme. Arch Biochem Biophys 342:92–98. doi:10.1006/abbi.1997.0115
Mutisya J, Sathish P, Sun C, Andersson L, Ahlandsberg S, Baguma Y, Palmqvist S, Odhiambo B, Aman P, Jansson C (2003) Starch branching enzymes in sorghum (Sorghum bicolor) and barley (Hordeum vulgare): comparative analyses of enzyme structure and gene expression. J Plant Physiol 160:921–930
Qin H, Zhou S, Y-z Zhang (2013) Characterization and expression analysis of starch branching enzymes in sweet potato. J Integr Agric 12:1530–1539. doi:10.1016/S2095-3119(13)60369-X
Boyer CD, Daniels RR, Shannon JC (1976) Abnormal starch granule formation in Zea mays L. endosperms possessing the amylose-extender mutant. Crop Sci 16:298–301
Yao Y, Thompson DB, Guiltinan MJ (2004) Maize starch-branching enzyme isoforms and amylopectin structure. In the absence of starch-branching enzyme IIb, the further absence of starch-branching enzyme Ia leads to increased branching. Plant Physiol 136:3515–3523. doi:10.1104/pp.104.043315
Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, Kosar-Hashemi B, Li Z, Rahman S, Morell M (2006) High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc Natl Acad Sci USA 103:3546–3551. doi:10.1073/pnas.0510737103
Schwall GP, Safford R, Westcott RJ, Jeffcoat R, Tayal A, Shi YC, Gidley MJ, Jobling SA (2000) Production of very-high-amylose potato starch by inhibition of SBE A and B. Nat Biotechnol 18:551–554. doi:10.1038/75427
Blauth SL, Kim KN, Klucinec J, Shannon JC, Thompson DB (2002) Identification of Mutator insertional mutants of starch-branching enzyme1 (sbe1) in Zea mays L. Plant Mol Biol 48:287–297
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373. doi:10.1093/nar/gkl198
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Wolfe KH, Gouy M, Yang YW, Sharp PM, Li WH (1989) Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA 86:6201–6205
Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW (2001) Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J 26:421–433
Theerawitaya C, Boriboonkaset T, Cha-Um S, Supaibulwatana K, Kirdmanee C (2012) Transcriptional regulations of the genes of starch metabolism and physiological changes in response to salt stress rice (Oryza sativa L.) seedlings. Physiol Mol Biol Plants 18:197–208. doi:10.1007/s12298-012-0114-x
Zhong LJ, Dong H, Cai XB, Feng YN, Ren P, Cheng FM (2012) Gene expression of the key enzymes controlling starch synthesis and metabolism in rice grain endosperm under effects of high temperature after anthesis. Ying Yong Sheng Tai Xue Bao 23:745–750
Acknowledgments
This study was supported by National Basic Research and Development Program (2010CB126600), China Agriculture Research System (CARS-12), and National international science and technology cooperation plan (2011DFB31690).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11010_2015_2445_MOESM2_ESM.jpg
Fig.S2 Expression profiles of MeSBE1 in leaves under various treatments. The relative expression variance was calculated as described in Methods and Materials, and cassava β-Actin gene was used to normalize the data as an internal control. Error bars were calculated based on four replicates. Different treatment and timing points were indicated in each table. Statistical difference (P<0.05) was indicated by different superscripts. A: MeSBE1 expression pattern in leaf under drought treatment. B: MeSBE1expression pattern in leaf under 200mM NaCl treatment. C: MeSBE1 expression pattern in leaf under high temperature treatment. D: MeSBE1expression pattern in leaf under cold treatment. E: MeSBE1expression pattern in leaf under 100uM abscisic acid (ABA) treatment. F: MeSBE1 expression pattern in leaf under 100uM jasmonic acid (JA) treatment. G: MeSBE1 expression pattern in leaf under 5mM salicylic acid (SA) treatment. Supplementary material 2 (JPG 583 kb)
11010_2015_2445_MOESM3_ESM.jpg
Fig.S3 Expression profiles of MeSBE2.1 in leaves under various treatments. The relative expression variance was calculated as described in Methods and Materials, and cassava β-Actin gene was used to normalize the data as an internal control. Error bars were calculated based on four replicates. Different treatment and timing points were indicated in each table. Statistical difference (P<0.05) was indicated by different superscripts. Supplementary material 3 (JPG 1437 kb)
11010_2015_2445_MOESM4_ESM.jpg
Fig.S4 Expression profiles of MeSBE3 in leaves under various treatments. The relative expression variance was calculated as described in Methods and Materials, and cassava β-Actin gene was used to normalize the data as an internal control. Error bars were calculated based on four replicates. Different treatment and timing points were indicated in each table. Statistical difference (P<0.05) was indicated by different superscripts. Supplementary material 4 (JPG 646 kb)
11010_2015_2445_MOESM5_ESM.jpg
Fig.S5 Expression profiles of MeSBE4 in leaves under various treatments. The relative expression variance was calculated as described in Methods and Materials, and cassava β-Actin gene was used to normalize the data as an internal control. Error bars were calculated based on four replicates. Different treatment and timing points were indicated in each table. Statistical difference (P<0.05) was indicated by different superscripts. Supplementary material 5 (JPG 634 kb)
11010_2015_2445_MOESM6_ESM.jpg
Fig.S6 Expression profiles of MeSBE5 in leaves under various treatments. The relative expression variance was calculated as described in Methods and Materials, and cassava β-Actin gene was used to normalize the data as an internal control. Error bars were calculated based on four replicates. Different treatment and timing points were indicated in each table. Statistical difference (P<0.05) was indicated by different superscripts. Supplementary material 6 (JPG 1477 kb)
Rights and permissions
About this article
Cite this article
Pei, J., Wang, H., Xia, Z. et al. Phylogeny and expression pattern of starch branching enzyme family genes in cassava (Manihot esculenta Crantz) under diverse environments. Mol Cell Biochem 406, 273–284 (2015). https://doi.org/10.1007/s11010-015-2445-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-015-2445-8