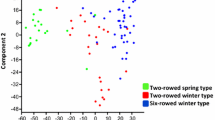
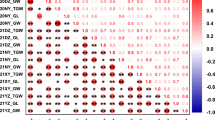
Abstract
Future barley cultivars will have to produce under the constraints of higher temperature in combination with increased concentrations of atmospheric carbon dioxide and ozone as a consequence of climate change. A diverse set of 167 spring barley genotypes was cultivated under elevated levels of temperature (+5 °C) and [CO2] (700 ppm) as single factors and in combination as well as under elevated [O3] (100–150 ppb) as single factor. The setting in general resembled changes projected by IPCC (AR5) to take place at the end of this century. A genome-wide association study (GWAS) was performed to identify markers for increased primary production under climate change conditions and reveal possible genes of interest. Phenotyped traits included grain yield, number of grains, number of ears per plant, aboveground vegetative biomass, harvest index and stability of the production parameters over the five applied treatments. The GWAS encompassed 7864 SNP markers (Illumina iselect), a compressed mixed linear model with the GAPIT package, and conservative validation of markers. A total of 60 marker-trait associations [−log10(P value) 2.97–5.58] were identified, e.g. grain yield under elevated temperature on barley chromosome 2H, static stability of grain yield on 7H, sites for exploitation of elevated [CO2] on 4H and 7H and associations under the two-factor treatment. Marker-trait associations identified from single-factor treatments were not retrieved, when elevated [CO2] and temperature were combined emphasizing the need for multifactor experiments. This GWA study identified markers and chromosome regions to be targeted in breeding for development of climate resilient cultivars.


Similar content being viewed by others
Abbreviations
- AllM:
-
Dataset of all markers
- BM:
-
Aboveground vegetative biomass
- EG:
-
Number of ears with grains
- ET:
-
Number of ears
- GN:
-
Number of grains
- GWAS:
-
Genome-wide association study
- GY:
-
Grain yield
- HI:
-
Harvest index
- LD:
-
Linkage disequilibrium
- MwP:
-
Markers from the AllM dataset with identified position
- QTL:
-
Quantitative trait loci
- SNP:
-
Single nucleotide polymorphism
References
Alpert M, Wanamaker S, Duma D et al (2011) A genome sequence resource for barley. Barley Genet Newsl 41:10–11
Backes G, Hatz B, Jahoor A, Fischbeck G (2003) RFLP diversity within and between major groups of barley in Europe. Plant Breed 122:291–299. doi:10.1046/j.1439-0523.2003.00810.x
Barrero-Sicilia C, Hernando-Amado S, González-Melendi P, Carbonero P (2011) Structure, expression profile and subcellular localisation of four different sucrose synthase genes from barley. Planta 234:391–403. doi:10.1007/s00425-011-1408-x
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
Bokszczanin KL, Fragkostefanakis S (2013) Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front Plant Sci 4:315. doi:10.3389/fpls.2013.00315
Bradbury PJ, Zhang Z, Kroon DE et al (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. doi:10.1093/bioinformatics/btm308
Brantestam AK, Bothmer R, Dayteg C et al (2007) Genetic diversity changes and relationships in spring barley (Hordeum vulgare L.) germplasm of Nordic and Baltic areas as shown by SSR markers. Genet Resour Crop Evol 54:749–758. doi:10.1007/s10722-006-9159-4
Bruhn D, Larsen KS, de Dato GD et al (2013) Improving the performance of infrared reflective night curtains for warming field plots. Agric For Meteorol 173:53–62. doi:10.1016/j.agrformet.2013.01.004
Cai S, Wu D, Jabeen Z et al (2013) Genome-wide association analysis of aluminum tolerance in cultivated and Tibetan wild barley. PLoS ONE 8:e69776. doi:10.1371/journal.pone.0069776
Caldwell KS, Russell J, Langridge P, Powell W (2006) Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare. Genetics 172:557–567. doi:10.1534/genetics.104.038489
Close TJ, Bhat PR, Lonardi S et al (2009) Development and implementation of high-throughput SNP genotyping in barley. BMC Genom 10:582. doi:10.1186/1471-2164-10-582
Collins M, Knutti R, Arblaster J, Dufresne J-L et al (2013) Long-term climate change: projections, commitments and irreversibility. In: Stocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM (eds.) Climate change 2013: the physical science basis. Contribution of working group I to the fifth assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge
Comadran J, Ramsay L, MacKenzie K et al (2011) Patterns of polymorphism and linkage disequilibrium in cultivated barley. Theor Appl Genet 122:523–531. doi:10.1007/s00122-010-1466-7
Comadran J, Kilian B, Russell J et al (2012) Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet 44:1388–1392. doi:10.1038/ng.2447
Ellermann T, Nøjgaard JK, Nordstrøm C, et al (2013) The Danish air quality monitoring programme. Annual summary for 2012. 59 pp
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
FAOSTAT (2014) Food supply, crops primary equivalent. http://faostat3.fao.org. Accessed Feb 2014
Franzaring J, Holz I, Fangmeier A (2013) Responses of old and modern cereals to CO2-fertilisation. 64:943–956. doi:10.1071/CP13311
Fuhrer J (2003) Agroecosystem responses to combinations of elevated CO2, ozone, and global climate change. Agric Ecosyst Environ 97:1–20. doi:10.1016/S0167-8809(03)00125-7
Hayes PM, Liu BH, Knapp SJ et al (1993) Quantitative trait locus effects and environmental interaction in a sample of North American barley germ plasm. Theor Appl Genet 87:392–401. doi:10.1007/BF01184929
Ingvordsen IPCC (2014) Summary for policymakers; climate change 2014: impacts, adaptation, and vulnerability. Contribution of working group II to the fifth assessment report of the Intergovernmental Panel on Climate Change 2014, pp 1–44
Ingvordsen CH, Backes G, Lyngkjær MF et al (2015) Significant decrease in yield under future climate conditions: stability and production of 138 spring barley accessions. Eur J Agron 63:105–113. doi:10.1016/j.eja.2014.12.003
Jannink J-L (2007) Identifying quantitative trait locus by genetic background interactions in association studies. Genetics 176:553–561. doi:10.1534/genetics.106.062992
Kang HM, Zaitlen NA, Wade CM et al (2008) Efficient control of population structure in model organism association mapping. Genetics 178:1709–1723. doi:10.1534/genetics.107.080101
Kimball B, Conley MM, Wang S et al (2007) Infrared heater arrays for warming ecosystem field plots. Glob Change Biol 14:309–320. doi:10.1111/j.1365-2486.2007.01486.x
Korte A, Farlow A (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9:29. doi:10.1186/1746-4811-9-29
Köszegi B, Karsai I, Mészáros K, Kovács G (2005) Study of the early growth and tillering capacity of barley (Hordeum vulgare L.) under changing temperature and light conditions. Acta Agron Hung 53:371–376
Kraakman ATW, Niks RE, Van den Berg PM et al (2004) Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics 168:435–446. doi:10.1534/genetics.104.026831
Lacaze X, Hayes PM, Korol A (2009) Genetics of phenotypic plasticity: QTL analysis in barley, Hordeum vulgare. Heredity 102:163–173. doi:10.1038/hdy.2008.76
Lipka AE, Tian F, Wang Q et al (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399. doi:10.1093/bioinformatics/bts444
Lobell DB, Schlenker W, Costa-Roberts J (2011) Climate trends and global crop production since 1980. Science 333:616–620. doi:10.1126/science.1204531
Long NV, Dolstra O, Malosetti M et al (2013) Association mapping of salt tolerance in barley (Hordeum vulgare L.). Theor Appl Genet Theor Angew Genet 126:2335–2351. doi:10.1007/s00122-013-2139-0
Manderscheid R, Weigel HJ (1997) Photosynthetic and growth responses of old and modern spring wheat cultivars to atmospheric CO2 enrichment. Agric Ecosyst Environ 64:65–73. doi:10.1016/S0167-8809(97)00020-0
Matthies IE, Sharma S, Weise S, Röder MS (2011) Sequence variation in the barley genes encoding sucrose synthase I and sucrose phosphate synthase II, and its association with variation in grain traits and malting quality. Euphytica 184:73–83. doi:10.1007/s10681-011-0563-2
Matthies IE, Hintum T, Weise S, Röder MS (2012) Population structure revealed by different marker types (SSR or DArT) has an impact on the results of genome-wide association mapping in European barley cultivars. Mol Breed 30:951–966. doi:10.1007/s11032-011-9678-3
Miralles DJ, Slafer GA (2007) Sink limitations to yield in wheat: How could it be reduced? J Agric Sci 145:139–149. doi:10.1017/S0021859607006752
Morison JIL, Lawlor DW (1999) Interactions between increasing CO2 concentration and temperature on plant growth. Plant Cell Environ 22:659–682
Nevo E, Fu Y-B, Pavlicek T et al (2012) Evolution of wild cereals during 28 years of global warming in Israel. Proc Natl Acad Sci USA 109:3412–3415. doi:10.1073/pnas.1121411109
Newton AC, Flavell AJ, George TS et al (2011) Crops that feed the world 4. Barley: a resilient crop? Strengths and weaknesses in the context of food security. Food Secur 3:141–178. doi:10.1007/s12571-011-0126-3
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rajala A, Hakala K, Mäkelä P, Peltonen-Sainio P (2011) Drought effect on grain number and grain weight at spike and spikelet level in six-row spring barley. J Agron Crop Sci 197:103–112. doi:10.1111/j.1439-037X.2010.00449.x
Roemer T (1917) Sind die ertragsreichen Sorten ertragssicherer? Mitteilung Deutsche Landwirtschafts-Gesellschaft 32:87–89
Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5:69–76. doi:10.1007/BF00020088
Roose JL, Wegener KM, Pakrasi HB (2007) The extrinsic proteins of photosystem II. Photosynth Res 92:369–387. doi:10.1007/s11120-006-9117-1
Rosenzweig C, Parry ML (1994) Potential impact of climate change on world food supply. Nature 367:133–138
Schweizer P, Stein N (2011) Large-scale data integration reveals colocalization of gene functional groups with meta-QTL for multiple disease resistance in barley. Mol Plant Microbe Interact 24:1492–1501. doi:10.1094/MPMI-05-11-0107
Stich B, Möhring J, Piepho H-P et al (2008) Comparison of mixed-model approaches for association mapping. Genetics 178:1745–1754. doi:10.1534/genetics.107.079707
Svenning J-C, Sandel B (2013) Disequilibrium vegetation dynamics under future climate change. Am J Bot 100:1266–1286. doi:10.3732/ajb.1200469
Tabangin ME, Woo JG, Martin LJ (2009) The effect of minor allele frequency on the likelihood of obtaining false positives. BMC Proc 3(Suppl 7):S41. doi:10.1186/1753-6561-3-S7-S41
R Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
The International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491:711–718. doi:10.1038/nature11543
Tondelli A, Xu X, Moragues M et al (2013) Structural and temporal variation in genetic diversity of european spring two-row barley cultivars and association mapping of quantitative traits. Plant Genome 6:1–14. doi:10.3835/plantgenome2013.03.0007
Turner A, Beales J, Faure S et al (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310:1031–1034. doi:10.1126/science.1117619
Ugarte C, Calderini DF, Slafer GA (2007) Grain weight and grain number responsiveness to pre-anthesis temperature in wheat, barley and triticale. Fields Crop Res 100:240–248. doi:10.1016/j.fcr.2006.07.010
UN (2012) World population prospects. United Nations, Department of Economic and Social Affairs Population Division. http://esa.un.org/wpp. Accessed 17 Jan 2014
Varshney RK, Paulo MJ, Grando S et al (2012) Genome wide association analyses for drought tolerance related traits in barley (Hordeum vulgare L.). Fields Crop Res 126:171–180. doi:10.1016/j.fcr.2011.10.008
Wang G, Schmalenbach I, von Korff M et al (2010) Association of barley photoperiod and vernalization genes with QTLs for flowering time and agronomic traits in a BC2DH population and a set of wild barley introgression lines. Theor Appl Genet 120:1559–1574. doi:10.1007/s00122-010-1276-y
Waugh R, Jannink J-L, Muehlbauer GJ, Ramsay L (2009) The emergence of whole genome association scans in barley. Curr Opin Plant Biol 12:218–222. doi:10.1016/j.pbi.2008.12.007
Wetterstrand K (2014) DNA sequencing costs: The NHGRI Genome Sequencing Program (GSP). http://www.genome.gov/sequencingcosts. Accessed 23 Jan 2014
Whitechurch EM, Slafer GA, Miralles DJ (2007) Variability in the duration of stem elongation in wheat and barley genotypes. J Agron Crop Sci 193:138–145. doi:10.1111/j.1439-037X.2007.00260.x
Wricke G (1962) Über eine Methode zur Erfassung der ökologischen Streubreite in Feldversuchen. Z Pflanzenzuchtg 47:92–96
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421
Zhang Z, Ersoz E, Lai C-Q et al (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42:355–360. doi:10.1038/ng.546
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome J 1:5. doi:10.3835/plantgenome2008.02.0089
Ziska LH, Morris CF, Goins EW (2004) Quantitative and qualitative evaluation of selected wheat varieties released since 1903 to increasing atmospheric carbon dioxide: Can yield sensitivity to carbon dioxide be a factor in wheat performance? Glob Change Biol 10:1810–1819. doi:10.1111/j.1365-2486.2004.00840.x
Acknowledgments
We thank all involved in the cultivation and harvest procedures and Allan Murphy and Esben Højrup for technical assistance in RERAF during the experiment. The accessions were provided by NordGen (the Nordic Genetic Resource Center; http://www.nordgen.org/), breeders of the Nordic network ‘Sustainable primary production in a changing climate’ (NordForsk) and a few came from the BAR-OF project (ICROFS, Denmark). The CO2 used in this study was generously supplied by Air Liquide Danmark A/S. The Nordic Council of Ministers supported the major part of this research through the network, ‘Sustainable primary production in a changing climate’. Other funders were The Danish Council for Independent Research: Technology and Production Sciences (FTP) via the project ‘Climate Change Effects on Plant Health’ (project 0602-00589B), and COBRA (Core Organic II).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ingvordsen, C.H., Backes, G., Lyngkjær, M.F. et al. Genome-wide association study of production and stability traits in barley cultivated under future climate scenarios. Mol Breeding 35, 84 (2015). https://doi.org/10.1007/s11032-015-0283-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0283-8