Abstract
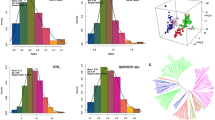
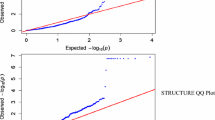
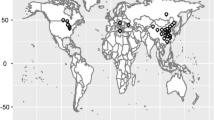
Symbiotic nitrogen fixation (SNF) in legume crops plays a crucial role in world sustainable agriculture. To analyze the genetic basis of SNF in soybean, a natural population composed of 267 typical germplasms genotyped with 5403 SNP markers was inoculated with CCBAU45436, an elite rhizobium strain from China. Eleven traits related to SNF were evaluated under greenhouse and field conditions, and a genome-wide association study (GWAS) was conducted. In total, 20 SNP loci (consisting of 38 SNP markers) that were significantly associated with 11 SNF-related characteristics were identified on eight soybean chromosomes. Three SNP loci located on chromosome Gm17 were associated with shoot nitrogen concentrations in the two environments. Two other SNP loci located on Gm17 in the adjacent region were associated with high nodule numbers and nodule fresh and dry weights. Moreover, the elite alleles and genotypes were selected based on the phenotypes of different groups, which further demonstrated the reliability of these associated SNP markers. Other colocalized SNP loci linked with different SNF traits were also detected. More importantly, some candidate genes found at the associated SNP loci showed different expression levels between the different developmental stages of soybean nodules, two of which were further verified by qRT-PCR results. These SNP loci and candidate genes could be applied in marker-assisted selection in breeding or map-based gene cloning in soybean for future genetic improvement.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Akond M, Liu SM, Kantartzi SK, Meksem K, Bellaloui N, Lightfoot DA, Yuan JZ, Wang DC, Anderson J, Kassem MA (2015) A SNP genetic linkage map based on the ‘Hamilton’ by ‘Spencer’ recombinant inbred line population identified QTL for seed isoflavone contents in soybean. Plant Breed 134(5):580–588
Bradbury PJ, Zhang ZW, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Broughton WJ, Dilworth MJ (1970) Plant nutrient solutions. In: Somasegaran P, Hoben HJ (eds) Methods in legume-rhizobium technology handbook for rhizobia NifTAL Project. Univ of Hawaii, Hawaii
Deshmukh R, Sonah H, Patil G, Chen W, Prince S, Mutava R, Vuong T, Valliyodan B, Nguyen HT (2014) Integrating omic approaches for abiotic stress tolerance in soybean. Front Plant Sci 5:1–14
Dhanapal AP, Ray JD, Singh SK, Hoyos-Villegas V, Smith JR, Purcell LC, Fritschi FB (2016) Genome-wide association mapping of soybean chlorophyll traits based on canopy spectral reflectance and leaf extracts. BMC Plant Biol 16(1):174
Dwivedi SL, Sahrawat KL, Upadhyaya HD, Mengoni A, Galardini M, Bazzicalupo M, Biondi EG, Hungria M, Kaschuk G, Blair MW, Ortiz R (2015) Advances in host plant and rhizobium genomics to enhance symbiotic nitrogen fixation in grain legumes. Adv Agron 129:1–116
Evanno GS, Regnaut SJ, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, Gresshoff PM (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52(1):61–76
Fritschi FB, Ray JD, Purcell LC, King CA, Smith JR, Charlson DV (2013) Diversity and implications of soybean stem nitrogen concentration. J Plant Nutr 36(14):2111–2131
Han YP, Zhao X, Cao GL, Wang Y, Li YH, Liu DY, Teng WL, Zhang ZW, Li DM, Qiu LJ, Zheng HK, Li WB (2015) Genetic characteristics of soybean resistance to HG type 0 and HG type 1.2.3.5.7 of the cyst nematode analyzed by genome-wide association mapping. BMC Genomics 16(1):598
Heilig JA, Beaver JS, Wright EM, Song QJ, Kelly JD (2017) QTL analysis of symbiotic nitrogen fixation in a black bean population. Crop Sci 57:118–129
Hwang S, King CA, Davies MK, Ray JD, Cregan PB, Purcell LC (2013) QTL analysis of shoot ureide and nitrogen concentrations in soybean [Glycine max (L.) Merr.]. Crop Sci 53(6):2421–2433
Hwang S, Ray JD, Cregan PB, King CA, Davies MK, Purcell LC (2014a) Genetics and mapping of quantitative traits for nodule number, weight, and size in soybean (Glycine max L. [Merr.]). Euphytica 195(3):419–434
Hwang EY, Song QJ, Jia GF, Specht JE, Hyten DL, Costa J, Cregan PB (2014b) A genome-wide association study of seed protein and oil content in soybean. BMC Genomics 15(1):1
Kamfwa K, Cichy KA, Kelly JD (2015) Genome-wide association analysis of symbiotic nitrogen fixation in common bean. Theor Appl Genet 128:1999–2017
Kong YB, Li XH, Wang B, Li WL, Du H, Zhang CY (2018) The soybean purple acid phosphatase GmPAP14 predominantly enhances external phytate utilization in plants. Front Plant Sci 9:292
Li CY, Chang HL, Wang JH, Zou JN, Shi Y, Zhang YJ, Wei W, Wang ZY, Wang JQ, Li QY, Zhu JY, Chen L, Li JY, Li SP, Liu XY, Jiang HW, Hu ZB, Liu CY, Yin ZG, Qi ZM, Zheng YL, Chen QS, Xin DW (2018a) Genetic analysis of nodule traits in soybean via wild soybean background population and high generation recombination inbred lines. Int J Agric Biol 20:2521–2528
Li XH, Tian R, Kamala S, Du H, Li WL, Kong YB, Zhang CY (2018b) Identification and verification of pleiotropic QTL controlling multiple amino acid contents in soybean seed. Euphytica 214:93
Li DM, Zhao X, Han YP, Li WB, Xie FT (2019) Genome-wide association mapping for seed protein and oil contents using a large panel of soybean accessions. Genomics 111(1):90–95
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129
Liu HY, Quampah A, Chen JH, Li JR, Huang ZR, He QL, Zhu SJ, Shi CH (2013) QTL mapping based on different genetic systems for essential amino acid contents in cottonseeds in different environments. PLoS One 8(3):e57531
Liu ZX, Li HH, Fan XH, Huang W, Yang JY, Li CD, Wen ZX, Li YH, Guan RX, Guo Y, Chang RZ, Wang DC, Wang SM, Qiu LJ (2016) Phenotypic characterization and genetic dissection of growth period traits in soybean (Glycine max) using association mapping. PLoS One 11(7):e0158602
Mastrodomenico AT, Purcell LC (2012) Soybean nitrogen fixation and nitrogen remobilization during reproductive development. Crop Sci 52(3):1281
Muñoz N, Qi X, Li MW, Xie M, Gao Y, Cheung MY, Wong FL, Lam HM (2016) Improvement in nitrogen fixation capacity could be part of the domestication process in soybean. Heredity 117(2):84–93
Nicolás MF, Hungria M, Arias CAA (2006) Identification of quantitative trait loci controlling nodulation and shoot mass in progenies from two Brazilian soybean cultivars. Field Crop Res 95:355–366
Ohlson EW, Seido SL, Mohammed S, Santos CAF, Timko MP (2018) QTL mapping of ineffective nodulation and nitrogen utilization-related traits in the IC-1 mutant of cowpea. Crop Sci 58:264–272
Peoples MB, Brockwell J, Herridge DF, Rochester IJ, Alves BJR, Urquiaga S, Boddey RM, Dakora FD, Bhattarai S, Maskey SL, Sampet C, Rerkasem B, Khan DF, Hauggaard-Nielsen H, Jensen ES (2009) The contributions of nitrogen-fixing crop legumes to the productivity of agricultural systems. Symbiosis 48:1–17
Ramaekers L, Galeano CH, Garzón N, Vanderleyden J, Blair MW (2013) Identifying quantitative trait loci for symbiotic nitrogen fixation capacity and related traits in common bean. Mol Breeding 31:163–180
Ramongolalaina C, Teraishi M, Okumoto Y (2018) QTLs underlying the genetic interrelationship between efficient compatibility of Bradyrhizobium strains with soybean and genistein secretion by soybean roots. PLoS One 13(4):e0194671
Ray JD, Dhanapal AP, Singh SK, Hoyos-Villegas V, Smith JR, Purcell LC, King CA, Boykin D, Cregan PB, Song QJ, Fritschi FB (2015) Genome-wide association study of ureide concentration in diverse maturity group IV soybean [Glycine max (L.) Merr.] accessions. G3-Genes Genom Genet 5:2391–2403
Santos MA, Nicolás MF, Hungria M (2006) Identification of QTL associated with the symbiosis of Bradyrhizobium japonicum, B. elkanii and soybean. Pesq Agropec Bras 41:67–75
Santos MA, Geraldi IO, Garcia AAF, Bortolatto N, Schiavon A, Hungria M (2013) Mapping of QTLs associated with biological nitrogen fixation traits in soybean. Hereditas 150:17–25
Sonah H, O’Donoughue L, Cober E, Rajcan I, Belzile F (2015) Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol J 13(2):211–221
Sun ZW, Wang XF, Liu ZW, Gu QS, Zhang Y, Li ZK, Ke HF, Yang J, Wu JH, Wu LQ, Zhang GY, Zhang CY, Ma ZY (2017) Genome-wide association study discovered genetic variation and candidate genes of fibre quality traits in Gossypium hirsutum L. Plant Biotechnol J 15(8):982–996
Tang Y, Liu XL, Wang JB, Li M, Wang QS, Tian F, Su ZB, Pan YC, Liu D, Lipka AE, Buckler ES, Zhang ZW (2016) GAPIT version 2: an enhanced integrated tool for genomic association and prediction. Plant Genome 9:1–9
Tanya P, Srinives P, Toojinda T, Vanavichit A, Lee SH (2005) Identification of SSR markers associated with N2-fixation components in soybean [Glycine max (L.) Merr.]. Korean J Genetics 27(4):351–359
Tian CF, Zhou YJ, Zhang YM, Li QQ, Zhang YZ, Li DF, Wang S, Wang J, Gilbert LB, Li YR, Chen WX (2012) Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc Natl Acad Sci U S A 109(22):8629–8634
Wang JH, Wang JQ, Liu CY, Ma C, Li CY, Zhang YQ, Qi ZM, Zhu RS, Shi Y, Zou JN, Li QY, Zhu JY, Wen YN, Sun ZJ, Liu HX, Jiang HW, Yin ZG, Hu ZB, Chen QS, Wu XX, Xin DW (2018) Identification of soybean genes whose expression is affected by the Ensifer fredii HH103 effector protein NopP. Int J Mol Sci 19(11):3438
Yan L, Hofmann N, Li SX, Ferreira ME, Song BH, Jiang GL, Ren SX, Quigley C, Fickus E, Cregan P, Song QJ (2017) Identification of QTL with large effect on seed weight in a selective population of soybean with genome-wide association and fixation index analyses. BMC Genomics 18(1):529
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA, a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82
Yuan SL, Li R, Chen SL, Chen HF, Zhang CJ, Chen LM, Hao QN, Shan ZH, Yang ZL, Qiu DZ, Zhang XJ, Zhou XN (2016) RNA-seq analysis of differential gene expression responding to different rhizobium strains in soybean (Glycine max) roots. Front Plant Sci 7:721
Zhang YF (2008) Screening of soybean and Rhizobial strains and mapping of genes for controlling nitrogen fixation in soybean cultivars at seedling stage. Thesis of Master, Shandong Normal University
Zhang H, Prithiviraj B, Charles TC, Driscoll BT, Smith DL (2003) Low temperature tolerant Bradyrhizobium japonicum strains allowing improved nodulation and nitrogen fixation of soybean in a short season (cool spring) area. Eur J Agrono 19(2):205–213
Zhang D, Song HN, Cheng H, Hao DR, Wang H, Kan GZ, Jin HX, Yu DY (2014) The acid phosphatase-encoding gene GmACP1 contributes to soybean tolerance to low-phosphorus stress. PLoS Genet 10(1):e1004061
Zhang J, Wen ZX, Li W, Zhang YW, Zhang LF, Dai HY, Wang DC, Xu R (2017) Genome-wide association study for soybean cyst nematode resistance in Chinese elite soybean cultivars. Mol Breeding 37(5):60
Zhang YJ, Liu XY, Chen L, Fu Y, Li CY, Qi ZM, Zou JN, Zhu RS, Li SP, Wei W, Wang JH, Chang HL, Shi Y, Wang JQ, Li QY, Zhu JY, Li JY, Jiang HW, Wu XX, Jia CG, Yin ZG, Hu ZB, Liu CY, Chen QS, Xin DW (2018a) Mining for genes encoding proteins associated with NopL of Sinorhizobium fredii HH103 using quantitative trait loci in soybean (Glycine max Merr.) recombinant inbred lines. Plant Soil 431(2):1–11
Zhang JP, Wang XZ, Lu YM, Bhusal SJ, Song QJ, Cregan PB, Yen Y, Brown M, Jiang GL (2018b) Genome-wide scan for seed composition provides insights into soybean quality improvement and the impacts of domestication and breeding. Mol Plant 11:460–472. https://doi.org/10.1016/j.molp.2017.12.016
Funding
This research was funded by the Project of Hebei Province Science and Technology Support Program (17927670H, 16227516D-1).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Huo, X., Li, X., Du, H. et al. Genetic loci and candidate genes of symbiotic nitrogen fixation–related characteristics revealed by a genome-wide association study in soybean. Mol Breeding 39, 127 (2019). https://doi.org/10.1007/s11032-019-1022-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-1022-3