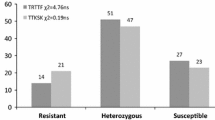
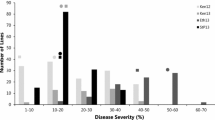
Abstract
Most wheat cultivars planted worldwide are susceptible to the stem rust Ug99 race group. To prepare for the potential spread of these races into South America, we aimed to identify genomic regions responsible for resistance to Ug99 race group in germplasm adapted to South America. Two RIL populations from a cross between a stem rust susceptible parent “Baguette 13” and resistant local parents “INIA Tero” and “BR23//CEP19/PF85490” were developed. Phenotypical evaluation was completed at the seedling stage in Uruguay and under field conditions in Uruguay and Kenya. Both RIL populations were genotyped using the GBS approach. Besides Sr24, three other resistance loci in “INIA Tero” were detected on chromosomes 2B, 6A, and 7B. All four QTL were effective to local races, whereas only the QTL on chromosome 2B was effective against the Ug99 race group. Besides Sr31, “BR23//CEP19/PF85490” also carries two other stem rust resistance loci on chromosomes 2B and 6A. All three explained the resistance in Uruguay, while only the QTL on 2B was effective to Ug99 in Kenya. The physical location suggested that the QTL identified on chromosome 2B in both populations may correspond to Sr28, which was confirmed using specific molecular markers. Further studies are needed to determine the relationship between QTL for resistance to local races identified on chromosomes 6A and 7B and previously reported resistance genes and QTL. The results of this study are highly relevant for breeding wheat cultivars with diverse and durable resistance to stem rust.




Similar content being viewed by others
References
Babiker EM, Gordon TC, Chao S, Rouse MN, Wanyera R, Newcomb M, Brown-Guedira G, Pretorius ZA, Bonman JM (2016) Genetic mapping of resistance to the Ug99 race group of Puccinia graminis f. sp. tritici in a spring wheat landrace CItr 4311. Theor Appl Genet 129:2161–2170. https://doi.org/10.1007/s00122-016-2764-5
Bajgain P, Rouse MN, Bulli P, Bhavani S, Gordon T, Wanyera R, Njau PN, Legesse W, Anderson JA, Pumphrey MO (2015) Association mapping of north American spring wheat breeding germplasm reveals loci conferring resistance to Ug99 and other African stem rust races. BMC Plant Biol 15:1–19. https://doi.org/10.1186/s12870-015-0628-9
Bariana HS, McIntosh RA (1993) Cytogenetic studies in wheat. XV. Location of rust resistance genes in VPM1 and their genetic linkage with other disease resistance genes in chromosome 2A. Genome 36:476–482. https://doi.org/10.1139/g93-065
Broman KW, Wu H, Sen Ś, Churchill GA (2003) R/QTL: QTL mapping in experimental crosses. Bioinformatics 19:889–890. https://doi.org/10.1093/bioinformatics/btg112
Castro M, Pereyra SA, Germán SE (2012) Resultados experimentales de la evaluación de cultivares de trigo período 2009–2010-2011. In: Resultados experimentales No 12. Convenio INASE-INIA, Colonia, Uruguay, p 24
Dreisigacker S, Sehgal D, Reyes JA et al (2017) CIMMYT wheat molecular genetics: laboratory protocols and applications to wheat breeding. CIMMYT, Mexico D.F
Dundas IS, Anugrahwati DR, Verlin DC, Park RF, Bariana HS, Mago R, Islam AKMR (2007) New sources of rust resistance from alien species: meliorating linked defects and discovery. Aust J Agric Res 58:545–549. https://doi.org/10.1071/AR07056
FAOSTAT (Food and Agriculture Organization of the United Nations) (2016) No Title. http://faostat.fao.org
Faris JD, Li WL, Liu DJ, Chen PD, Gill BS (1999) Candidate gene analysis of quantitative disease resistance in wheat. Theor Appl Genet 98:219–225. https://doi.org/10.1007/s001220051061
Gao L, Rouse MN, Mihalyov PD, Bulli P, Pumphrey MO, Anderson JA (2017) Genetic characterization of stem rust resistance in a global spring wheat germplasm collection. Crop Sci 57:2575–2589. https://doi.org/10.2135/cropsci2017.03.0159
Germán S, Barcellos A, Chaves M, Kohli M, Campos P, de Viedma L (2007) The situation of common wheat rusts in the southern cone of America and perspectives for control. Aust J Agric Res 58:620. https://doi.org/10.1071/ar06149
Germán SE, Chaves M, Campos P et al (2009) Borlaug global rust initiative (2009, Ciudad Obregón, Sonora, MX). Proceedings: oral papers and posters. In: McIntosh R (ed) Are rust pathogens under control in the southern cone of South America? BGRI, Sonora, pp 75–64
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9:e90346. https://doi.org/10.1371/journal.pone.0090346
Haile JK, Nachit MM, Hammer K, Badebo A, Röder MS (2012) QTL mapping of resistance to race Ug99 of Puccinia graminis f. sp. tritici in durum wheat (Triticum durum Desf.). Mol Breed 30:1479–1493. https://doi.org/10.1007/s11032-012-9734-7
Hibert CW, Rouse MN, Nirmala J, Fetch T (2017) Genetic mapping of stem rust resistance to Puccinia graminis f.sp tritici race TRTTF in the Canadian wheat cultivar harvest. Phytopath 107(2):192–197. https://doi.org/10.1094/PHYTO-05-16-0186-R
Hiebert CW, Fetch TG, Zegeye T (2010) Genetics and mapping of stem rust resistance to Ug99 in the wheat cultivar Webster. Theor Appl Genet 121:65–69. https://doi.org/10.1007/s00122-010-1291-z
International Wheat Genome Sequencing Consortium (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345(80):1251788. https://doi.org/10.1126/science.1251788
Jin Y, Singh RP (2006) Resistance in U.S. wheat to recent eastern African isolates of Puccinia graminis f. sp. tritici with virulence to resistance gene Sr31. Plant Dis 90:476–480. https://doi.org/10.1094/PD-90-0476
Jin Y, Singh RP, Ward RW, Wanyera R, Kinyua M, Njau P, Fetch T, Pretorius ZA, Yahyaoui A (2007) Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinia graminis f. sp. tritici. Plant Dis 91:1096–1099. https://doi.org/10.1094/pdis-91-9-1096
Jin Y, Szabo LJ, Pretorius ZA, Singh RP, Ward R, Fetch T Jr (2008) Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp. tritici. Plant Dis 92:923–926. https://doi.org/10.1094/PDIS-92-6-0923
Jin Y, Szabo LJ, Rouse MN, Fetch T Jr, Pretorius ZA, Wanyera R, Njau P (2009) Detection of virulence to resistance gene Sr36 within the TTKS race lineage of Puccinia graminis f. sp. tritici. Plant Dis 93:367–370. https://doi.org/10.1094/pdis-93-4-0367
Kaur J, Bansal UK, Khanna R et al (2009) Molecular mapping of stem rust resistance in HD2009 / WL711 recombinant inbred line population. Int J Plant Breed 3:28–33
Klindworth DL, Niu Z, Chao S et al (2012) Introgression and characterization of a Goatgrass gene for a high level of resistance to Ug99 stem rust in Tetraploid wheat. 2:665–673. https://doi.org/10.1534/g3.112.002386
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugenics 12:172–175. https://doi.org/10.1111/j.1469-1809.1943.tb02321.x
Li H, Singh S, Bhavani S, Singh RP, Sehgal D, Basnet BR, Vikram P, Burgueno-Ferreira J, Huerta-Espino J (2016) Identification of genomic associations for adult plant resistance in the background of popular south Asian wheat cultivar, PBW343. Front Plant Sci 7. https://doi.org/10.3389/fpls.2016.01674
Lopez-Vera EE, Nelson S, Singh RP, Basnet BR, Haley SD, Bhavani S, Huerta-Espino J, Xoconostle-Cazares BG, Ruiz-Medrano R, Rouse MN, Singh S (2014) Resistance to stem rust Ug99 in six bread wheat cultivars maps to chromosome 6DS. Theor Appl Genet 127:231–239. https://doi.org/10.1007/s00122-013-2212-8
Maccaferri M, Sanguineti MC, Mantovani P, Demontis A, Massi A, Ammar K, Kolmer JA, Czembor JH, Ezrati S, Tuberosa R (2010) Association mapping of leaf rust response in durum wheat. Mol Breed 26:189–228. https://doi.org/10.1007/s11032-009-9353-0
Mcintosh RA, Luig NH, Baker EP (1967) Genetic and Cytogenetic Studies of Stem Rust, Leaf Rust, and Powdery Mildew Resistances in Hope and Related Wheat Cultivars. Australian Journal of Biological Sciences 20(6):1181
Mago R, Spielmeyer W, Lawrence GJ, Lagudah E, Ellis J, Pryor A (2002) Identification and mapping of molecular markers linked to rust resistance genes located on chromosome 1RS of rye using wheat-rye translocation lines. Theor Appl Genet 104:1317–1324. https://doi.org/10.1007/s00122-002-0879-3
Mago R, Bariana HS, Dundas IS, Spielmeyer W (2005) Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor Appl Genet 111:496–504. https://doi.org/10.1007/s00122-005-2039-z
Mago R, Zhang P, Bariana HS, Verlin DC, Bansal UK, Ellis JG, Dundas IS (2009) Development of wheat lines carrying stem rust resistance gene Sr39 with reduced Aegilops speltoides chromatin and simple PCR markers for marker-assisted selection. Theor Appl Genet 119:1441–1450. https://doi.org/10.1007/s00122-009-1146-7
McIntosh RA (1972) Cytogenetical studies in wheat VI. Chromosome location and linkage studies involving Sr13 and Sr8 for reaction to Puccinia graminis f. sp. Tritici. Aust J Biol Sci 25:765–773. https://doi.org/10.1071/BI9720765
McIntosh RA (1978) Cytogenetical studies in wheat. Heredity (Edinb) 41:71–82. https://doi.org/10.1038/hdy.1978.65
McIntosh RA, Wellings CR, Park RF (1995) Wheat rusts: an atlas of resistance genes. Australia CSIRO, Melburne
McIntosh RA, Yamazaki Y, Dubcovsky J et al (2014) 12th international wheat genetics symposium. In: Catalogue of gene symbols for wheat 2013(14 supplement)
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283. https://doi.org/10.1016/j.cj.2015.01.001
Nazari K, Mafi M, Yahyaoui A, Singh RP, Park RF (2009) Detection of wheat stem rust (Puccinia graminis f. sp. tritici) race TTKSK (Ug99) in Iran. Plant Dis 93:317–317. https://doi.org/10.1094/pdis-93-3-0317b
Njau P, Bhavani S, Huerta-Espino J et al (2013) Identification of QTL associated with durable adult plant resistance to stem rust race Ug99 in wheat cultivar ‘Pavon 76.’. Euphytica 190:33–44. https://doi.org/10.1007/s10681-012-0763-4
Peterson R, Campbell A, Hannah A (1948) A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can J Res 26:496–500. https://doi.org/10.1139/cjr48c-033
Poland JA, Brown PJ, Sorrells ME, Jannink JL (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7:e32253. https://doi.org/10.1371/journal.pone.0032253
Pretorius ZA, Singh RP, Wagoire WW, Payne TS (2000) Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f.sp. tritici in Uganda. Plant Dis 82:203–203. https://doi.org/10.1094/PDIS.2000.84.2.203B
Pretorius ZA, Bender CM, Visser B, Terefe T (2010) First report of a Puccinia graminis f. sp. tritici race virulent to the Sr24 and Sr31 wheat stem rust resistance genes in South Africa. Plant Dis 94:784–784. https://doi.org/10.1094/PDIS-94-6-0784C
Qi LL, Pumphrey MO, Friebe B, Zhang P, Qian C, Bowden RL, Rouse MN, Jin Y, Gill BS (2011) A novel Robertsonian translocation event leads to transfer of a stem rust resistance gene (Sr52) effective against race Ug99 from Dasypyrum villosum into bread wheat. Theor Appl Genet 123:159–167. https://doi.org/10.1007/s00122-011-1574-z
R Development Core Team (2014) R: A language and environment for statistical computing
Roelfs A, Broers L, Dubin H et al (1992) Las Royas del Trigo. CIMMYT, Mexico D.F
Rosas JE, Martínez S, Blanco P, Pérez de Vida F, Bonnecarrère V, Mosquera G, Cruz M, Garaycochea S, Monteverde E, McCouch S, Germán S, Jannink JL, Gutiérrez L (2017) Resistance to multiple temperate and tropical stem and sheath diseases of Rice. Plant Genome 11:1–13. https://doi.org/10.3835/plantgenome2017.03.0029
Rouse MN, Nava IC, Chao S, Anderson JA, Jin Y (2012) Identification of markers linked to the race Ug99 effective stem rust resistance gene Sr28 in wheat (Triticum aestivum L.). Theor Appl Genet 125:877–885. https://doi.org/10.1007/s00122-012-1879-6
Rouse MN, Nirmala J, Jin Y, Chao S, Fetch TG Jr, Pretorius ZA, Hiebert CW (2014) Characterization of Sr9h, a wheat stem rust resistance allele effective to Ug99. Theor Appl Genet 127:1681–1688. https://doi.org/10.1007/s00122-014-2330-y
Simón MR, Worland AJ, Struik PC (2004) Influence of plant height and heading date on the expression of the resistance to Septoria tritici blotch in near isogenic lines of wheat. Crop Sci 44:2078–2085. https://doi.org/10.2135/cropsci2004.2078
Singh RP, Rajaran S (1991) Resistance to Puccinia recondita f. sp. tritici in 50 Mexican bread wheat cultivars. Crop Sci 31:1472–1479. https://doi.org/10.2135/cropsci1991.0011183x003100060016x
Singh RP, Huerta-Espino J, William HM (2005) Genetics and breeding for durable resistance to leaf and stripe rusts in wheat. Turkish J Agric For 29:121–127. https://doi.org/10.3906/tar-0402-2
Singh RP, Hodson DP, Jin Y et al (2006) Current status, likely migration and strategies to mitigate the threat to wheat production from race Ug99 (TTKS) of stem rust pathogen. CAB Rev Perspect Agric Vet Sci Nutr Nat Resour 1:1–13. https://doi.org/10.1079/pavsnnr20061054
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, Njau P, Herrera-Foessel S, Singh PK, Singh S, Govindan V (2011a) The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol 49:465–481. https://doi.org/10.1146/annurev-phyto-072910-095423
Singh RP, Huerta-Espino J, Bhavani S, Herrera-Foessel SA, Singh D, Singh PK, Velu G, Mason RE, Jin Y, Njau P, Crossa J (2011b) Race non-specific resistance to rust diseases in CIMMYT spring wheats. Euphytica 179:175–186. https://doi.org/10.1007/s10681-010-0322-9
Singh S, Singh RP, Bhavani S, Huerta-Espino J, Eugenio LVE (2013) QTL mapping of slow-rusting, adult plant resistance to race Ug99 of stem rust fungus in PBW343/Muu RIL population. Theor Appl Genet 126:1367–1375. https://doi.org/10.1007/s00122-013-2058-0
Singh RP, Hodson DP, Jin Y, Lagudah ES, Ayliffe MA, Bhavani S, Rouse MN, Pretorius ZA, Szabo LJ, Huerta-Espino J, Basnet BR, Lan C, Hovmøller MS (2015) Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology 105:872–884. https://doi.org/10.1094/PHYTO-01-15-0030-FI
Stakman EC, Stewart DM, Loegering WQ (1962) Identification of physiologic races of Puccinia graminis var. tritici. USDA, Washington
Stubbs RW, Prescott JM (1986) Manual De Metodologia Sobre Las Enfermedades De Los Cereales. CIMMYT, Mexico D.F
Tsilo TJ, Jin Y, Anderson JA (2007) Microsatellite markers linked to stem rust resistance allele Sr9a in wheat. Crop Sci 47:2013–2020. https://doi.org/10.2135/cropsci2007.02.0087
Wu S, Pumphrey M, Bai G (2009) Molecular mapping of stem-rust-resistance gene Sr40 in wheat. Crop Sci 49:1681–1686. https://doi.org/10.2135/cropsci2008.11.0666
Yu LX, Barbier H, Rouse MN, Singh S, Singh RP, Bhavani S, Huerta-Espino J, Sorrells ME (2014) A consensus map for Ug99 stem rust resistance loci in wheat. Theor Appl Genet 127:1561–1581. https://doi.org/10.1007/s00122-014-2326-7
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421
Acknowledgments
We thank Fernando Pereira and Noelia Pérez (Instituto Nacional de Investigación Agropecuaria—INIA Uruguay) for assistance with field and greenhouse phenotyping. We also thank Ruth Wanyera for assistance with field phenotyping in Kenya. This research was supported by the INNOVAGRO program of the National Agency of Research and Innovation of Uruguay (ANII), through the project FSA_1_2013_1_12980.
Author information
Authors and Affiliations
Contributions
SG, MQ, AC, and LG designed the hypothesis and experiments. SG created the populations. SBaraibar, RG, and SG conducted disease phenotyping in Uruguay and MR and SBhavani in Kenya. SBaraibar, BL, and PS conducted statistical analysis. SBaraibar conducted laboratory analysis. SBaraibar and SG wrote the paper. All authors contributed to the discussion of the results and edited the paper.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Baraibar, S., García, R., Silva, P. et al. QTL mapping of resistance to Ug99 and other stem rust pathogen races in bread wheat. Mol Breeding 40, 82 (2020). https://doi.org/10.1007/s11032-020-01153-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-020-01153-5