Abstract
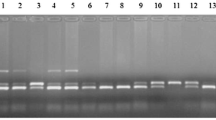
Bovine mastitis is a very complex and common disease of dairy cattle and a major source of economic losses to the dairy industry worldwide. In this study, the bovine breast cancer 1, early onset gene (BRCA1) was taken as a candidate gene for mastitis resistance. The main object of this study was to investigate whether the BRCA1 gene was associated with mastitis in cattle. Through DNA sequencing, Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) and Created Restriction Site PCR (CRS-PCR) methods, three SNPs (G22231T, T25025A, and C28300A) were detected and twenty-four combinations of these SNPs were observed. The single SNP and their genetic effects on somatic cell score (SCS) were evaluated and a significant association with SCS was found in C28300A. The mean of genotype EE was significantly lower than those of genotypes EF and FF. The results of combined genotypes analysis of three SNPs showed that BBDDFF genotype with the highest SCS were easily for the mastitis susceptibility, whereas AACCEE genotype with the lowest SCS were favorable for the mastitis resistance. The information provided in the present study will be very useful for improving mastitis resistance in dairy cattle by marker-assisted selection.
Similar content being viewed by others
References
Janzen JJ (1970) Economic losses resulting from mastitis. Rev J Dairy Sci 53:1151–1161
Lescourret F, Coulon JB (1994) Modeling the impact of mastitis on milk production by dairy cows. J Dairy Sci 77:2289–2301
Nash DL, Rogers GW, Cooper JB, Hargrove GL, Keown JF (2003) Heritability of intramammary infections at first parturition and relationships with sire transmitting abilities for somatic cell score, udder type traits, productive life, and protein yield. J Dairy Sci 86:2684–2695
Ruegg PL (2003) Investigation of mastitis problems on farms. Vet Clin N Am Food A 19:47–73
Zhang LP, Gan QF, Ma TH, Li HD, Wang XP, Li JY, Gao X, Chen JB, Ren HY, Xu SZ (2009) Toll-like receptor 2 gene polymorphism and its relationship with SCS in dairy cattle. Anim Biotechnol 20:87–95
Chu MX, Ye SC, Qiao L, Wang JX, Feng T, Huang DW, Cao GL, Di R, Fang L, Chen GH (2011) Polymorphism of exon 2 of BoLA-DRB3 gene and its relationship with somatic cell score in Beijing Holstein cows. Mol Biol Rep. doi:10.1007/s11033-011-1052-3
Chen R, Yang Z, Ji D, Mao Y, Chen Y, Li Y, Wu H, Wang X, Chang L (2011) Polymorphisms of the IL8 gene correlate with milking traits, SCS and mRNA level in Chinese Holstein. Mol Biol Rep 38:4083–4088
Yuan ZR, Li J, Liu L, Zhang LP, Zhang LM, Chen C, Chen XJ, Gao X, Li JY, Chen JB, Gao HJ, Xu SZ (2011) Single nucleotide polymorphism of CACNA2D1 gene and its association with milk somatic cell score in cattle. Mol Biol Rep 38:5179–5183
Rupp R, Boichard D (1999) Genetic parameters for clinical mastitis, somatic cell score, production, udder type traits, and milking ease in first lactation Holsteins. J Dairy Sci 82:2198–2204
Lund MS, Jensen J, Petersen PH (1999) Estimation of genetic and phenotypic parameters for clinical mastitis, somatic cell production deviance, and protein yield in dairy cattle using Gibbs sampling. J Dairy Sci 82:1045–1051
Carlen E, Strandberg E, Roth A (2004) Genetic parameters for clinical mastitis, somatic cell score, and production in the first three lactations of Swedish Holstein cows. J Dairy Sci 87:3062–3070
Heringstad B, Gianola D, Chang YM, Odegard J, Klemetsdal G (2006) Genetic associations between clinical mastitis and somatic cell score in early first-lactation cows. J Dairy Sci 89:2236–2244
Emanuelson U, Danell B, Philipsson J (1988) Genetic parameters for clinical mastitis, somatic cell counts, and milk production estimated by multiple-trait restricted maximum likelihood. J Dairy Sci 71:467–476
Shook GE (2006) Major advances in determining appropriate selection goals. J Dairy Sci 89:1349–1361
Schutz MM (1994) Genetic evaluation of somatic cell scores for United States dairy cattle. J Dairy Sci 77:2113–2129
Philipsson J, Ral G, Berglund B (1995) Somatic-cell count as a selection criterion for mastitis resistance in dairy-cattle. Livest Prod Sci 41:195–200
Shook GE, Schutz MM (1994) Selection on somatic cell score to improve resistance to mastitis in the United States. J Dairy Sci 77:648–658
Hou GY, Yuan ZR, Gao X, Li JY, Gao HJ, Chen JB, Xu SZ (2010) Genetic polymorphisms of the CACNA2D1 gene and their association with carcass and meat quality traits in cattle. Biochem Genet 48:751–759
Liu HY, Tian WQ, Zan LS, Wang HB, Cui HA (2010) Mutations of MC4R gene and its association with economic traits in Qinchuan cattle. Mol Biol Rep 37:535–540
Zhang CL, Wang YH, Chen H, Lan XY, Lei CZ, Fang XT (2009) Association between variants in the 5′-untranslated region of the bovine MC4R gene and two growth traits in Nanyang cattle. Mol Biol Rep 36:1839–1843
te Pas MF, Soumillion A, Harders FL, Verburg FJ, van den Bosch TJ, Galesloot P, Meuwissen TH (1999) Influences of myogenin genotypes on birth weight, growth rate, carcass weight, backfat thickness, and lean weight of pigs. J Anim Sci 77:2352–2356
Zhou G, Dudgeon C, Li M, Cao Y, Zhang L, Jin H (2010) Molecular cloning of the HGD gene and association of SNPs with meat quality traits in Chinese red cattle. Mol Biol Rep 37:603–611
Fontanesi L, Scotti E, Buttazzoni L, Dall’Olio S, Bagnato A, Lo Fiego DP, Davoli R, Russo V (2010) Confirmed association between a single nucleotide polymorphism in the FTO gene and obesity-related traits in heavy pigs. Mol Biol Rep 37:461–466
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W et al (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66–71
Xu J, Wang B, Zhang Y, Li R, Wang Y and Zhang S (2011) Clinical implications for BRCA gene mutation in breast cancer. Mol Biol Rep DOI 10.1007/s11033-011-1073-y
Krum SA, Womack JE, Lane TF (2003) Bovine BRCA1 shows classic responses to genotoxic stress but low in vitro transcriptional activation activity. Oncogene 22:6032–6044
Kennedy RD, Quinn JE, Johnston PG, Harkin DP (2002) BRCA1: mechanisms of inactivation and implications for management of patients. Lancet 360:1007–1014
Antoniou AC, Gayther SA, Stratton JF, Ponder BA, Easton DF (2000) Risk models for familial ovarian and breast cancer. Genet Epidemiol 18:173–190
Wang F, Fang Q, Ge Z, Yu N, Xu S and Fan X (2011) Common BRCA1 and BRCA2 mutations in breast cancer families: a meta-analysis from systematic review. Mol Biol Rep. doi:10.1007/s11033-011-0958-0
Mahfoudh W, Bouaouina N, Ahmed SB, Gabbouj S, Shan J, Mathew R, Uhrhammer N, Bignon YJ, Troudi W, Elgaaied AB, Hassen E, Chouchane L (2012) Hereditary breast cancer in Middle Eastern and North African (MENA) populations: identification of novel, recurrent and founder BRCA1 mutations in the Tunisian population. Mol Biol Rep 39:1037–1046
Narod SA, Ford D, Devilee P, Barkardottir RB, Lynch HT, Smith SA, Ponder BA, Weber BL, Garber JE, Birch JM et al (1995) An evaluation of genetic heterogeneity in 145 breast-ovarian cancer families. Breast cancer linkage consortium. Am J Hum Genet 56:254–264
Neuhausen SL (1999) Ethnic differences in cancer risk resulting from genetic variation. Cancer 86:2575–2582
Rebbeck TR (1999) Inherited genetic predisposition in breast cancer. A population-based perspective. Cancer 86:2493–2501
Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, Timmerman MM, Brody LC, Tucker MA (1997) The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. New Engl J Med 336:1401–1408
Lane TF, Deng C, Elson A, Lyu MS, Kozak CA, Leder P (1995) Expression of BRCA1 is associated with terminal differentiation of ectodermally and mesodermally derived tissues in mice. Genes Dev 9:2712–2722
Yang YP, Womack JE (1998) Parallel radiation hybrid mapping: a powerful tool for high-resolution genomic comparison. Genome Res 8:731–736
Bennewitz J, Reinsch N, Grohs C, Leveziel H, Malafosse A, Thomsen H, Xu N, Looft C, Kuhn C, Brockmann GA, Schwerin M, Weimann C, Hiendleder S, Erhardt G, Medjugorac I, Russ I, Förster M, Brenig B, Reinhardt F, Reents R, Averdunk G, Blümel J, Boichard D, Kalm E (2003) Combined analysis of data from two granddaughter designs: a simple strategy for QTL confirmation and increasing experimental power in dairy cattle. Genet Sel Evol 35:319–338
Bennewitz J, Reinsch N, Guiard V, Fritz S, Thomsen H, Looft C, Kuhn C, Schwerin M, Weimann C, Erhardt G, Reinhardt F, Reents R, Boichard D, Kalm E (2004) Multiple quantitative trait loci mapping with cofactors and application of alternative variants of the false discovery rate in an enlarged granddaughter design. Genetics 168:1019–1027
Daetwyler HD, Schenkel FS, Sargolzaei M, Robinson JA (2008) A genome scan to detect quantitative trait loci for economically important traits in Holstein cattle using two methods and a dense single nucleotide polymorphism map. J Dairy Sci 91:3225–3236
Mullenbach R, Lagoda PJ, Welter C (1989) An efficient salt-chloroform extraction of DNA from blood and tissues. Trends Genet 5:391
Zhao CJ, Li N, Deng XM (2003) The establishment of method for identifying SNP genotype by CRS-PCR. Yi Chuan 25:327–329
Yeh FC, Yang RC, Boyle T (1999) POPGENE Version 1.31, Microsoft window-based freeware for population genetic analysis. University of Alberta and Centre for International Forestry Research, Canada
Zheng X, Ju Z, Wang J, Li Q, Huang J, Zhang A, Zhong J, Wang C (2011) Single nucleotide polymorphisms, haplotypes and combined genotypes of LAP3 gene in bovine and their association with milk production traits. Mol Biol Rep 38:4053–4061
Wang X, Xu S, Gao X, Ren H, Chen J (2007) Genetic polymorphism of TLR4 gene and correlation with mastitis in cattle. J Genet Genomics 34:406–412
Nott A, Meislin SH, Moore MJ (2003) A quantitative analysis of intron effects on mammalian gene expression. RNA 9:607–617
Fallin D, Cohen A, Essioux L, Chumakov I, Blumenfeld M, Cohen D, Schork NJ (2001) Genetic analysis of case/control data using estimated haplotype frequencies: application to APOE locus variation and Alzheimer’s disease. Genome Res 11:143–151
Liu ZH, Chen SL (2010) ER regulates an evolutionarily conserved apoptosis pathway. Biochem Biophys Res Commun 400(1):34–38
Liu ZH, Sun X (2008) Coronavirus phylogeny based on base-base correlation. Int J Bioinform Res Appl 4(2):211–220
Liu ZH, Meng J, Sun X (2008) A novel feature-based method for whole genome phylogenetic analysis without alignment: application to HEV genotyping and subtyping. Biochem Biophys Res Commun 368(2):223–230
Liu ZH, Jiao D, Sun X (2005) Classifying genomic sequences by sequence feature analysis. Genomics Proteomics Bioinformatics 3(4):201–205
Acknowledgment
The study was financially supported by the New Variety of Transgenic Organism Great Breeding Program (NO.2008ZX08007-002 and NO.2009ZX08009-157B) and the earmarked fund for Modern Agro-industry Technology Research System (No.CARS-38); National Natural Science Foundation of China (81102746), Beijing Natural Science Foundation (5113033), Scientific Research Foundation of the State Human Resource Ministry and the Education Ministry for Returned Chinese Scholars, New Star Project of Peking Union Medical College, Youth Foundation of Peking Union Medical College, the Research Fund for the Doctoral Program of Higher Education (20111106120028), “Major Drug Discovery” major science and technology research “12nd Five-Year Plan” (2012ZX09301-002-001) China Medical Board of New York (A2009001) granted to Zhihua Liu.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Z. Yuan, G. Chu and Y. Dan have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yuan, Z., Chu, G., Dan, Y. et al. BRCA1: a new candidate gene for bovine mastitis and its association analysis between single nucleotide polymorphisms and milk somatic cell score. Mol Biol Rep 39, 6625–6631 (2012). https://doi.org/10.1007/s11033-012-1467-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1467-5