Abstract
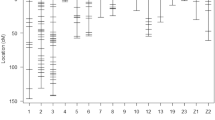
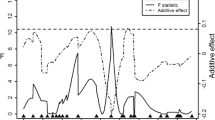
Japanese quail is an animal model in biological studies and also a commercial bird for eggs and meat production. This study was conducted to map quantitative trait loci (QTL) affecting live weight in Japanese quail. An F2 mapping population was developed by crossing two diverse lines (meat type and egg layer) of Japanese quail. A total number of 34 F1 and 422 F2 progeny were produced by reciprocal crossing of eight pairs of parental birds. All the birds from three generations were genotyped for SSR markers that were spread across all the autosomal linkage groups. The studied traits were hatching weight and live weights at 1–5 weeks of age. QTL analysis was conducted by the regression interval mapping. Significant QTL were detected on chromosomes 1, 2, 3 (chromosome-wide significant) and 5 (genome-wide significant, P < 0.05) for body weight. Although the additive effect of the detected QTL on chromosome 5 was significant, the dominance and imprinting effects were not significant. This finding is the first report of a genome-wide significant QTL associated with live weight in Japanese quail. Our results point out to candidate DNA regions affecting live weight, a trait of great economic relevance to the Japanese quail breeding. Although these results enhance our current knowledge about the genetic control of live weight in the Japanese quail, it should be noted that the initial QTL results from the experimental designs such as backcross or F2 cannot be applied directly to the breeding programs and require further validation within the commercial lines.




Similar content being viewed by others
References
Jacobsson L, Park H, Wahlberg P, Fredriksson R, Perez-Enciso M, Siegel P, Andersson L (2005) Many QTLs with minor additive effects are associated with a large difference in growth between two selection lines in chickens. Genet Res 86:115–125
Esmailizadeh AK (2010) A partial genome scan to identify quantitative trait loci affecting birthweight in kermani sheep. Small Rumin Res 94:73–78
Groenen MA, Cheng HH, Bumstead N (2000) A consensus linkage map of the chicken genome. Genome Res 10:137–147
Kayang BB, Vignal A, Inoue-Murayama M, Miwa M, Monvosin JL, Ito S, Minvieele F (2004) A first-generation microsatellite linkage map of the Japanese quail. Anim Genet 35:195–200
Minvielle F, Kayang BB, Inoue-Murayama M, Miwa M, Vignal A, Gourichon D, Neau A, Monvoisin JL, Ito S (2005) Microsatellite mapping of QTL affecting growth, feed consumption, egg production, tonic immobility and body temperature of Japanese quail. BMC Genom 6:87
Esmailizadeh AK, Baghizadeh A, Ahmadizadeh M (2012) Genetic mapping of quantitative trait loci affecting bodyweight on chromosome 1 in a commercial strain of Japanese quail. Anim Prod Sci 52:64–68
Seaton G, Hernandez J, Grunchec JA, White I, Allen J, de Koning DJ, Wei W, Berry D, Halley C, Knott SA (2006) GridQTL: a grid portal for QTL mapping of compute intensive datasets. In: Proceedings of 8th world congress on genetics applied to livestock production, Belo Horizonte, 13–18 August 2006
Carlborg O, Kerje S, Schutz K, Jacobsson L, Jensen P, Andersson L (2003) A global search reveals epistatic interaction between QTL for early growth in the chicken. Genome Res 13:413–421
Le Rouzic A, Alvarez-Castro JM, Carlborg O (2008) Dissection of the genetic architecture of body weight in chicken reveals the impact of epistasis on domestication traits. Genetics 179:1591–1599
Shibusawa M, Minai S, Nishida-Umehara C, Suzuki T, Mano T, Yamada K, Namikawa T, Matsuda Y (2001) A comparative cytogenetic study of chromosome homology between chicken and Japanese quail. Cytogenet Cell Genet 95(1–2):103–109
Wang SZ, Hu XX, Wang ZP, Li XC, Wang QG, Wang YX, Tang ZQ, Li H (2012) Quantitative trait loci associated with body weight and abdominal fat traits on chicken chromosomes 3, 5 and 7. Genet Mol Res 11(2):956–965
Li H, Deeb N, Zhou H, Mitchell AD, Ashwell CM, Lamont SJ (2003) Chicken quantitative trait loci for growth and body composition associated with transforming growth factor-beta genes. Poult Sci 82:347–356
Zhou H, Deeb N, Evock-Clover CM, Ashwell CM, Lamont SJ (2006) Genome-wide linkage analysis to identify chromosomal regions affecting phenotypic traits in the chicken: growth and average daily gain. Poult Sci 85:1700–1711
Nie QH, Fang MX, Xie L, Shen X, Liu J, Luo ZP, Shi JJ, Zhang XQ (2010) Associations of ATGL gene polymorphisms with chicken growth and fat traits. J Appl Genet 51:185–191
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Iranmanesh, M., Esmailizadeh, A., Mohammad Abadi, M.R. et al. A molecular genome scan to identify DNA segments associated with live weight in Japanese quail. Mol Biol Rep 43, 1267–1272 (2016). https://doi.org/10.1007/s11033-016-4059-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-4059-y