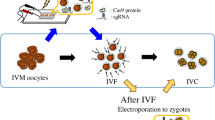
Abstract
The CRISPR/Cas9 system now allows for unprecedented possibilities of genome editing. However, there are some limitations, including achieving efficient one-step multiple genome targeting to save costs, time, and ensure high quality. In the present study, we investigated the efficiency of one-step multiple gene modification by electroporation in porcine zygotes using pooled guide RNAs (gRNAs) targeting CMAH, GHR, GGTA1, and PDX1. We first selected the best-performing gRNA from three different designs for each gene based on the effect on embryo development and mutation efficiency. The three gRNAs showed equivalent effects on the rates of blastocyst formation in each targeted gene; however, gRNAs CMAH #2, GHR #3, GGTA1 #3, and PDX1 #3 showed the highest biallelic mutation rate, although the total mutation rate of PDX1 #3 was significantly lower than that of PDX1 #1. Therefore, CMAH #2, GHR #3, GGTA1 #3, and PDX1 #1 were used as a mixture in electroporation to further clarify whether multiple genes can be targeted simultaneously. Individual sequencing of 43 blastocysts at the target sites of each gene showed mutations in one and two target genes in twenty-four (55.8%) and nine (20.9%) blastocysts, respectively. No mutation was detected in any target gene in ten (23.3%) blastocysts and no blastocysts had a mutation in three or more target genes. These results indicate that electroporation could effectively deliver multiple gRNAs and Cas9 protein into porcine zygotes to target multiple genes in a one-step process. However, the technique requires further development to increase the success rate of multiple gene modification.


Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are included in the manuscript.
References
Brinkman EK, Chen T, Amendola M, van Steensel B (2014) Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42:e168
Cao J, Wu L, Zhang S-M, Lu M, Cheung WK, Cai W, Gale M, Xu Q, Yan Q (2016) An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res 44:e149–e149
Chen S-J (2019) Minimizing off-target effects in CRISPR-Cas9 genome editing. Springer, New York
Cradick TJ, Fine EJ, Antico CJ, Bao G (2013) CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 41:9584–9592
Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE (2014) Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat Biotechnol 32:1262
Fischer K, Rieblinger B, Hein R, Sfriso R, Zuber J, Fischer A, Klinger B, Liang W, Flisikowski K, Kurome M, Zakhartchenko V, Kessler B, Wolf E, Rieben R, Schwinzer R, Kind A, Schnieke A (2020) Viable pigs after simultaneous inactivation of porcine MHC class I and three xenoreactive antigen genes GGTA1, CMAH and B4GALNT2. Xenotransplantation 27:e12560
Graf R, Li X, Chu VT, Rajewsky K (2019) sgRNA sequence motifs blocking efficient CRISPR/Cas9-mediated gene editing. Cell Rep 26(1098–1103):e1093
Han X, Liu Z, Chan JM, Zhang K, Li Y, Zeng Z, Li N, Zu Y, Qin L (2015) CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci Adv 1:e1500454
Hinrichs A, Kessler B, Kurome M, Blutke A, Kemter E, Bernau M, Scholz AM, Rathkolb B, Renner S, Bultmann S, Leonhardt H, de Angelis MH, Nagashima H, Hoeflich A, Blum WF, Bidlingmaier M, Wanke R, Dahlhoff M, Wolf E (2018) Growth hormone receptor-deficient pigs resemble the pathophysiology of human Laron syndrome and reveal altered activation of signaling cascades in the liver. Mol Metab 11:113–128
Hirata M, Wittayarat M, Hirano T, Nguyen NT, Le QA, Namula Z, Fahrudin M, Tanihara F, Otoi T (2019) The Relationship between Embryonic Development and the Efficiency of Target Mutations in Porcine Endogenous Retroviruses (PERVs) Pol Genes in Porcine Embryos. Animals 9:593
Hurh S, Kang B, Choi I, Cho B, Lee EM, Kim H, Kim YJ, Chung YS, Jeong JC, Hwang JI, Kim JY, Lee BC, Surh CD, Yang J, Ahn C (2016) Human antibody reactivity against xenogeneic N-glycolylneuraminic acid and galactose-alpha-1,3-galactose antigen. Xenotransplantation 23:279–292
Iqbal K, Barg-Kues B, Broll S, Bode J, Niemann H, Kues WA (2009) Cytoplasmic injection of circular plasmids allows targeted expression in mammalian embryos. Biotechniques 47:959–968
Kaneko T, Sakuma T, Yamamoto T, Mashimo T (2014) Simple knockout by electroporation of engineered endonucleases into intact rat embryos. Sci Rep 4:6382
Martens GR, Reyes LM, Li P, Butler JR, Ladowski JM, Estrada JL, Sidner RA, Eckhoff DE, Tector M, Tector AJ (2017) Humoral reactivity of renal transplant-waitlisted patients to cells from GGTA1/CMAH/B4GalNT2, and SLA class I knockout pigs. Transplantation 101:e86–e92
Matsunari H, Nagashima H, Watanabe M, Umeyama K, Nakano K, Nagaya M, Kobayashi T, Yamaguchi T, Sumazaki R, Herzenberg LA, Nakauchi H (2013) Blastocyst complementation generates exogenic pancreas in vivo in apancreatic cloned pigs. Proc Natl Acad Sci USA 110:4557–4562
Namula Z, Wittayarat M, Hirata M, Hirano T, Nguyen NT, Le QA, Fahrudin M, Tanihara F, Otoi T (2019) Genome mutation after the introduction of the gene editing by electroporation of Cas9 protein (GEEP) system into bovine putative zygotes. Vitro Cell Dev Biol Anim 55:598–603
Nguyen DH, Tangvoranuntakul P, Varki A (2005) Effects of natural human antibodies against a nonhuman sialic acid that metabolically incorporates into activated and malignant immune cells. J Immunol 175:228–236
Nishio K, Tanihara F, Nguyen TV, Kunihara T, Nii M, Hirata M, Takemoto T, Otoi T (2018) Effects of voltage strength during electroporation on the development and quality of in vitro-produced porcine embryos. Reprod Domest Anim 53:313–318
Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W (2014) Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156:836–843
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183
Ren X, Yang Z, Xu J, Sun J, Mao D, Hu Y, Yang S-J, Qiao H-H, Wang X, Hu Q (2014) Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell reports 9:1151–1162
Sekine R, Kawata T, Muramoto T (2018) CRISPR/Cas9 mediated targeting of multiple genes in Dictyostelium. Sci Rep 8:8471
Sharei A, Zoldan J, Adamo A, Sim WY, Cho N, Jackson E, Mao S, Schneider S, Han M-J, Lytton-Jean A (2013) A vector-free microfluidic platform for intracellular delivery. Proc Natl Acad Sci 110:2082–2087
Song J, Yang D, Ruan J, Zhang J, Chen YE, Xu J (2017) Production of immunodeficient rabbits by multiplex embryo transfer and multiplex gene targeting. Sci Rep 7:12202
Tanihara F, Hirata M, Nguyen NT, Le QA, Wittayarat M, Fahrudin M, Hirano T, Otoi T (2019) Generation of CD163-edited pig via electroporation of the CRISPR/Cas9 system into porcine in vitro-fertilized zygotes. Anim Biotechnol 111:1–8
Tanihara F, Takemoto T, Kitagawa E, Rao S, Do LTK, Onishi A, Yamashita Y, Kosugi C, Suzuki H, Sembon S (2016) Somatic cell reprogramming-free generation of genetically modified pigs. Sci Adv 2:e1600803
Valenti MT, Serena M, Carbonare LD, Zipeto D (2019) CRISPR/Cas system: an emerging technology in stem cell research. World J Stem Cells 11:937–956
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918
Zhou J, Shen B, Zhang W, Wang J, Yang J, Chen L, Zhang N, Zhu K, Xu J, Hu B (2014) One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol 46:49–55
Acknowledgements
We thank the Nippon Food Packer, K. K. Shikoku (Tokushima, Japan) for supplying the pig ovaries. This study was supported in part by the Program of Open Innovation Platform with Enterprises, Research Institute and Academia (OPERA) Grant Number JPMJOP1613 from the Japan Science and Technology Agency (JST), and KAKENHI Grant Numbers JP17H03938, JP18K12062 and JP19K16014 from the Japan Society for the Promotion of Science (JSPS). We acknowledge Tokushima University for their financial support of the Research Clusters program of Tokushima University (No. 1701001).
Author information
Authors and Affiliations
Contributions
MH, FT, and TO conceived the study and wrote the manuscript. MH, FT, and ZN performed the majority of experiments. TO designed the study, coordinated all of the experiments, and reviewed the manuscript. QAL, QL, NTN, and KT contributed to the laboratory work and statistical analysis. MW and YS revised the manuscript. All of the authors read and accepted the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics approval
All procedures involving the handling and collection of materials from animals were approved by the Animal Research Committee of Tokushima University (Approval Number: T28-21), and were performed in accordance with the Guidelines for Animal Experiments of Tokushima University based on the rules of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) JAPAN, and the Ministry of the Environment.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hirata, M., Wittayarat, M., Namula, Z. et al. Evaluation of multiple gene targeting in porcine embryos by the CRISPR/Cas9 system using electroporation. Mol Biol Rep 47, 5073–5079 (2020). https://doi.org/10.1007/s11033-020-05576-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05576-3