Abstract
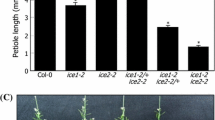
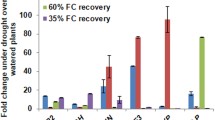
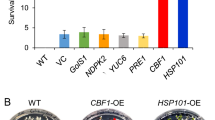
C-Repeat Binding Factors (CBFs) are DNA-binding transcriptional activators of gene pathways imparting freezing tolerance. Poaceae contain three CBF subfamilies, two of which, HvCBF3/CBFIII and HvCBF4/CBFIV, are unique to this taxon. To gain mechanistic insight into HvCBF4/CBFIV CBFs we overexpressed Hv-CBF2A in spring barley (Hordeum vulgare) cultivar ‘Golden Promise’. The Hv-CBF2A overexpressing lines exhibited stunted growth, poor yield, and greater freezing tolerance compared to non-transformed ‘Golden Promise’. Differences in freezing tolerance were apparent only upon cold acclimation. During cold acclimation freezing tolerance of the Hv-CBF2A overexpressing lines increased more rapidly than that of ‘Golden Promise’ and paralleled the freezing tolerance of the winter hardy barley ‘Dicktoo’. Transcript levels of candidate CBF target genes, COR14B and DHN5 were increased in the overexpressor lines at warm temperatures, and at cold temperatures they accumulated to much higher levels in the Hv-CBF2A overexpressors than in ‘Golden Promise’. Hv-CBF2A overexpression also increased transcript levels of other CBF genes at FROST RESISTANCE-H2-H2 (FR-H2) possessing CRT/DRE sites in their upstream regions, the most notable of which was CBF12. CBF12 transcript levels exhibited a relatively constant incremental increase above levels in ‘Golden Promise’ both at warm and cold. These data indicate that Hv-CBF2A activates target genes at warm temperatures and that transcript accumulation for some of these targets is greatly enhanced by cold temperatures.






Similar content being viewed by others
References
Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20:2117–2129
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidmen JG, Smith JA et al (1993) Current protocols in molecular biology. Greene Publishing Associates/Wiley, NY
Badawi M, Danyluk J, Boucho B, Houde M, Sarhan F (2007) The CBF gene family in hexaploid wheat and its relationship to the phylogenetic complexity of cereal CBFs. Mol Genet Genomics 277:533–554
Båga M, Chodaparambil SV, Limin AE, Pecar M, Fowler DB, Chibbar RN (2007) Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Funct Integr Genomics 7:53–68
Baneyx F, Mujacic M (2004) Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol 22:1399–1408
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57:289–300
Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB (2004) The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol 134:224–236
Campoli C, Matus-Cadiz MA, Pozniak CJ, Cattivelli L, Fowler DB (2009) Comparative expression of Cbf genes in the Triticeae under different acclimation induction temperatures. Mol Genet Genomics 282:141–152
Cattivelli L, Baldi P, Crosatti C, Di Fonzo N, Faccioli P, Grossi M et al (2002) Chromosome regions and stress-related sequences involved in resistance to abiotic stress in Triticeae. Plant Mol Biol 48:649–665
Chang Y, von Zitzewitz J, Hayes PM, Chen THH (2003) High frequency plant regeneration from immature embryos of an elite barley cultivar (Hordeum vulgare L. cv. Morex). Plant Cell Rep 21:733–738
Choi DW, Zhu B, Close TJ (1999) The barley (Hordeum vulgare L.) dehydrin multigene family: sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theor Appl Genet 98:1234–1247
Dal Bosco C, Busconi M, Govoni C, Baldi P, Stanca AM, Crosatti C et al (2003) cor gene expression in barley mutants affected in chloroplast development and photosynthetic electron transport. Plant Physiol 131:793–802
Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB et al (2009) A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci USA 106:5996–6001
Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Toth B et al (2004) Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’ (winter) × ’Tremois’ (spring) barley map. Theor Appl Genet 108:670–680
Francia E, Barabaschi D, Tondelli A, Laido G, Rizza F, Stanca AM et al (2007) Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor Appl Genet 115:1083–1091
Gill G, Ptashne M (1988) Negative effect of the transcriptional activator GAL4. Nature 334:721–724
Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16:433–442
Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124:1854–1865
Gilmour SJ, Fowler SG, Thomashow MF (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54:767–781
Gusta LV, O’Connor BJ, Gao YP, Jana S (2001) A re-evaluation of controlled freeze-tests and controlled environment hardening conditions to estimate the winter survival potential of hardy winter wheats. Can J Plant Sci 81:241–246
Horstmann V, Huether CM, Jost W, Reski R, Decker EL (2004) Quantitative promoter analysis in Physcomitrella patens: a set of plant vectors activating gene expression within three orders of magnitude. BMC Biotechnol 4:13
Horvath H, Huang J, Wong OT, von Wettstein D (2002) Experiences with genetic transformation of barley and characteristics of transgenic plants. In: Slafer GA, Molina-Cano JL, Savin R, Araus JL, Romagosa I (eds) Barley science: recent advances from molecular biology to agronomy of yield and quality. Food Products Press, Binghamton, pp 143–176
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M et al (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153
Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ et al (2001) Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127:910–917
Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280:104–106
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287–291
Knox AK, Li C, Vagujfalvi A, Galiba G, Stockinger EJ, Dubcovsky J (2008) Identification of candidate CBF genes for the frost tolerance locus Fr-A m 2 in Triticum monococcum. Plant Mol Biol 67:257–270
Knox AK, Dhillon T, Cheng H, Tondelli A, Pecchioni N, Stockinger EJ (2010) CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet 121:21–35
Koag MC, Wilkens S, Fenton RD, Resnik J, Vo E, Close TJ (2009) The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol 150:1503–1514
Kobayashi F, Takumi S, Kume S, Ishibashi M, Ohno R, Murai K et al (2005) Regulation by Vrn-1/Fr-1 chromosomal intervals of CBF-mediated Cor/Lea gene expression and freezing tolerance in common wheat. J Exp Bot 56:887–895
Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Bio/Technology 9:963–967
Levine M, Manley JL (1989) Transcriptional repression of eukaryotic promoters. Cell 59:405–408
Limin AE, Fowler DB (2006) Low-temperature tolerance and genetic potential in wheat (Triticum aestivum L.): response to photoperiod, vernalization, and plant development. Planta 224:360–366
Limin A, Corey A, Hayes P, Fowler DB (2007) Low-temperature acclimation of barley cultivars used as parents in mapping populations: response to photoperiod, vernalization and phenological development. Planta 226:139–146
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K et al (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Lourenco T, Saibo N, Batista R, Ricardo CP, Oliveira MM (2011) Inducible and constitutive expression of HvCBF4 in rice leads to differential gene expression and drought tolerance. Biol Plant 55:653–663
Miller AK, Galiba G, Dubcovsky J (2006) A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-A m2 in Triticum monococcum. Mol Genet Genomics 275:193–203
Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A et al (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J 9:230–249
Oh SJ, Kwon CW, Choi DW, Song SI, Kim JK (2007) Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol J 5:646–656
Olien CR (1964) Freezing processes in the crown of ‘Hudson’ barley, Hordeum vulgare (L., emend. Lam.) Hudson. Crop Sci 4:91–95
Pellegrineschi A, Reynolds M, Pacheco M, Brito RM, Almeraya R, Yamaguchi-Shinozaki K et al (2004) Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 47:493–500
Pino MT, Skinner JS, Park EJ, Jeknić Z, Hayes PM, Thomashow MF et al (2007) Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol J 5:591–604
Sanders PR, Winter JA, Barnason AR, Rogers SG, Fraley RT (1987) Comparison of cauliflower mosaic virus 35S and nopaline synthase promoters in transgenic plants. Nucleic Acids Res 15:1543–1558
Skinner JS, von Zitzewitz J, Szucs P, Marquez-Cedillo L, Filichkin T, Amundsen K et al (2005) Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol Biol 59:533–551
Skinner JS, Szucs P, von Zitzewitz J, Marquez-Cedillo L, Filichkin T, Stockinger EJ et al (2006) Mapping of barley homologs to genes that regulate low temperature tolerance in Arabidopsis. Theor Appl Genet 112:832–842
Soltész A, Smedley M, Vashegyi I, Galiba G, Harwood W, Vágújfalvi A (2013) Transgenic barley lines prove the involvement of TaCBF14 and TaCBF15 in the cold acclimation process and in frost tolerance. J Exp Bot 64:1849–1862
Stockinger EJ, Mulinix CA, Long CM, Brettin TS, Iezzoni AF (1996) A linkage map of sweet cherry based on RAPD analysis of a microspore-derived callus culture population. J Hered 87:214–218
Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94:1035–1040
Stockinger EJ, Skinner JS, Gardner KG, Francia E, Pecchioni N (2007) Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J 51:308–321
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Vágújfalvi A, Crosatti C, Galiba G, Dubcovsky J, Cattivelli L (2000) Two loci on wheat chromosome 5A regulate the differential cold-dependent expression of the cor14b gene in frost-tolerant and frost-sensitive genotypes. Mol Gen Genet 263:194–200
Vágújfalvi A, Galiba G, Cattivelli L, Dubcovsky J (2003) The cold-regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Mol Genet Genomics 269:60–67
van Buskirk HA, Thomashow MF (2006) Arabidopsis transcription factors regulating cold acclimation. Physiol Plant 126:72–80
Veisz O, Sutka J (1989) The relationships of hardening period and the expression of frost resistance in chromosome substitution lines of wheat. Euphytica 43:41–45
Vera A, Gonzalez-Montalban N, Aris A, Villaverde A (2007) The conformational quality of insoluble recombinant proteins is enhanced at low growth temperatures. Biotechnol Bioeng 96:1101–1106
Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41:195–211
Walkerpeach CR, Velten J (1994) Agrobacterium-mediated gene transfer to plant cells: cointegrate and binary vector systems. In: Gelvin SB, Schilperoort RA (eds) Plant molecular biology manual, vol B1, 2nd edn. Kluwer Academic, Dordrecht, pp 1–19
Wang MB, Li ZY, Matthews PR, Upadhyaya NM, Waterhouse PM (1998) Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. Acta Hortic 461:401–407
Wang Z, Triezenberg SJ, Thomashow MF, Stockinger EJ (2005) Multiple hydrophobic motifs in Arabidopsis CBF1 COOH-terminus provide functional redundancy in trans-activation. Plant Mol Biol 58:543–559
Xue GP (2003) The DNA-binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature. Plant J 33:373–383
Zarka DG, Vogel JT, Cook D, Thomashow MF (2003) Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol 133:910–918
Acknowledgments
This research was supported in part by grants from the NSF Plant Genome Project (DBI 0110124 and DBI 0701709). Katherine Pillman was supported by a fellowship from the Australian Centre for Plant Functional Genomics (Adelaide, Australia). Alfonso Cuesta-Marcos was supported by a postdoctoral fellowship from the Spanish Ministerio de Ciencia e Innovación (MICINN). Salaries and research support in the Stockinger lab provided by state and federal funds appropriated to The Ohio State University, Ohio Agricultural Research and Development Center, the Ohio Plant Biotechnology Consortium, and USDA-CSREES subaward CO396A-F. We thank Dr. Wang Ming-Bo (CSIRO, Australia) for providing pWBVec10a binary vector and Dr. Neil Shirley (The University of Adelaide, Australia) for providing assistance with qRT-PCR. We also thank Dr. Michael F. Thomashow for helpful suggestions and Drs. David Mackey and Esther van der Knaap for critically reviewing the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Zoran Jeknić, Katherine A. Pillman, and Taniya Dhillon have contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jeknić, Z., Pillman, K.A., Dhillon, T. et al. Hv-CBF2A overexpression in barley accelerates COR gene transcript accumulation and acquisition of freezing tolerance during cold acclimation. Plant Mol Biol 84, 67–82 (2014). https://doi.org/10.1007/s11103-013-0119-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0119-z