Abstract
Key message
Salt stress induces the degradation of 14-3-3 proteins, and affects the localization of 14-3-3 λ. Both the modulation of 14-3-3 protein stability and the subcellular localization of these proteins are involved in salt tolerance in plants.
Abstract
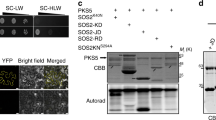
Salt tolerance in plants is regulated by multiple signaling pathways, including the salt overly sensitive (SOS) pathway, of which the SOS2 protein is a key component. SOS2 is activated under salt stress to enhance salt tolerance in plants. We previously identified 14-3-3 λ and κ as important regulators of salt tolerance. Both proteins interact with SOS2 to inhibit its kinase activity under normal growth conditions. In response to salt stress, 14-3-3 proteins dissociate from SOS2, releasing its activity and activating the SOS pathway to confer salt tolerance (Zhou et al. Plant Cell 26:1166–1182, 2014). Here we report that salt stress promotes the degradation of 14-3-3 λ and κ, at least in part via the actions of SOS3-like calcium binding protein 8/calcineurin-B-like10, and also decreases the plasma membrane (PM) localization of 14-3-3 λ. Salt stress also partially represses the interaction of SOS2 and 14-3-3 λ at the PM, but activates PM-localized SOS2. Together, these results suggest that, in plants, both the modulation of 14-3-3 stability and the subcellular localization of these proteins in response to salt stress are important for SOS2 activation and salt tolerance. These data provide new insights into the biological roles of 14-3-3 proteins in modulating salt tolerance.





Similar content being viewed by others
References
Albrecht V, Ritz O, Linder S, Harter K, Kudla J (2001) The NAF domain defines a novel protein–protein interaction module conserved in Ca2+–regulated kinases. EMBO J 20:1051–1063
Batistic, O, Kudla, J (2009). Plant calcineurin B-like proteins and their interacting protein kinases. Biochim Biophys Acta 1793:985–992
Batistič O, Sorek N, Schültke S, Yalovsky S, Kudla J (2008) Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20:1346–1362
Chen D, Dai D, Hua Y, Qi W (2015) p53 suppresses 14-3-3 γ by stimulating proteasome-mediated 14-3-3 γ protein degradation. Int J Oncol 46(2):818–824
Denison FC, Paul A-L, Zupanska AK, Ferl RJ (2011) 14-3-3 proteins in plant physiology. Semin Cell Dev Biol 22:720–727
Du W, Lin H, Chen S, Wu Y, Zhang J, Fuglsang AT, Palmgren MG, Wu W, Guo Y (2011) Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in Arabidopsis. Plant Physiol 156:2235–2243
Guo Y, Halfter U, Ishitani M, Zhu J-K (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13:1383–1400
Haglund K, Dikic I (2005) Ubiquitylation and cell signaling. EMBO J 24:3353–3359
Halfter U, Ishitani M, Zhu J-K (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97:3735–3740
Hochstrasser M (1996) Ubiquitin-dependent protein degradation. Annu Rev Genet 30:405–439
Ishitani M, Liu J, Halfter U, Kim C-S, Shi W, Zhu J-K (2000) SOS3 function in plant salt tolerance requiresN-myristoylation and calcium binding. Plant Cell 12:1667–1678
Kang HK, Nam KH (2016) Reverse function of ROS-induced CBL10 during salt and drought stress responses. Plant Sci 243:49–55
Katiyar-Agarwal S, Zhu J, Kim K, Agarwal M, Fu X, Huang A, Zhu J-K (2006) The plasma membrane Na+/H+antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 103:18816–18821
Kim BG, Waadt R, Cheong YH, Pandey GK, Dominguez-Solis JR, Schültke S, Lee SC, Kudla J, Luan S (2007) The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J 52:473–484
Kim WY, Ali Z, Park HJ, Park SJ, Cha JY, Perez-Hormaeche J et al (2013) Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat Commun 4(1):273–275
Lin H, Yang Y, Quan R, Mendoza I, Wu Y, Du W, Zhao S, Schumaker KS, Pardo JM, Guo Y (2009) Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21:1607–1619
Liu J, Zhu J-K (1998) A calcium sensor homolog required for plant salt tolerance. Science 280:1943–1945
Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97:3730–3734
Meng L, Wang Y, Yao S (2015) Arabidopsis AINTEGUMENTA mediates salt tolerance by trans-repressing SCABP8. J Cell Sci 128(15):2919–2927
Oecking C, Jaspert N (2009) Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr Opin Plant Biol 12:760–765
Ohta M, Guo Y, Halfter U, Zhu J-K (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2 C ABI2. Proc Natl Acad Sci USA 100:11771–11776
Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K (2002) Regulation of SOS1, a plasma membrane Na+/H+exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99:8436–8441
Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y (2007) SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis hoots from salt stress. Plant Cell 19:1415–1431
Quintero FJ, Ohta M, Shi H, Zhu J-K, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+homeostasis. Proc Natl Acad Sci USA 99:9061–9066
Quintero FJ, Martinez-Atienza J, Villalta I, Jiang X, Kim W-Y, Ali Z, Fujii H, Mendoza I, Yun D-J, Zhu J-K, Pardo JM (2011) Activation of the plasma membrane Na+/H+antiporter Salt-Overly Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc Natl Acad Sci USA 108:2611–2616
Sato T, Maekawa S, Yasuda S, Domeki Y, Sueyoshi K, Fujiwara M, Fukao Y, Goto DB, Yamaguchi J (2011) Identification of 14-3-3 proteins as a target of ATL31 ubiquitin ligase, a regulator of the C/N response in Arabidopsis. Plant J 68(1):137–146
Sheen J (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127:1466–1475
Shi H, Ishitani M, Kim C, Zhu J-K (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+antiporter. Proc Natl Acad Sci USA 97:6896–6901
Shi H, Quintero FJ, Pardo JM, Zhu J-K (2002) The putative plasma membrane Na+/H+antiporter SOS1 controls long distance Na+transport in plants. Plant Cell 14:465–477
Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan L-M, Deng XW (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21:2378–2390
Weinl S, Kudla J (2009) The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184:517–528
Welchman RL, Gordon C, Mayer RJ (2005) Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol 6:599–609
Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y, Zhang W (2010) Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol 188:762–773
Zhou H, Lin H, Chen S, Becker K, Yang Y, Zhao J, Kudla J, Schumaker KS, Guo Y (2014) Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26:1166–1182
Zhu J-K, Xiong L, Ishitani M, Liu J, Lee H, Stevenson B, Shi W (1998) Identification of genes important for environmental stress tolerance in plants. In: Breeding and biotechnology of environmental stress in rice, Sato Y (ed) (Sapporo: Hokkaido National Agricultural Experiment Station) pp 105–113
Acknowledgments
We thank Dr. Honghui Lin, Dr. Dehui Xi, and Dr. Dawei Zhang from Sichuan University for their critical reading of the manuscript and their stimulating discussions; and Chongwu Wang, Changxi Chen, Yuan Xue, and Jianfang Li from China Agricultural University for their excellent technical assistance. This work was supported by the Sichuan University Start-Up Funding to H.Z.
Author contributions
H.Z. and Y.G. designed the research. T.T., J.C., E.Z., and H.Z. performed most of the research. T.T. and H.Z. analyzed the data. J.Z. and Y.Y. performed the research on the analysis of cytosol/PM isolation. Y.G. and H.Z. contributed to the discussion and wrote the article.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tan, T., Cai, J., Zhan, E. et al. Stability and localization of 14-3-3 proteins are involved in salt tolerance in Arabidopsis . Plant Mol Biol 92, 391–400 (2016). https://doi.org/10.1007/s11103-016-0520-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0520-5