Abstract
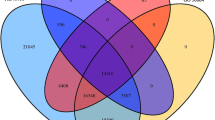
Soil salinity is a major abiotic stress that affects crop productivity. Garden asparagus (Asparagus officinalis L.) is a perennial plant with some salt tolerance. However, little is known about its response mechanism to salinity stress. In this study, we conducted transcriptome analysis in the leaves of A. officinalis seedlings treated with NaCl solution for 0 h, 1 h, 24 h, and 72 h using the Illumina HiSeq™ 2500 sequencing platform. Compared with the control (0 h), 1027, 3387, and 3358 differentially expressed genes (DEGs) were identified at 1 h, 24 h, and 72 h, respectively. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses revealed that these DEGs were highly enriched in carbon metabolism, ion transport, and reactive oxygen species (ROS) metabolism functional categories, suggesting their key positions in salinity stress responses. Moreover, k-means clustering categorized these DEGs into five kinds of expression patterns at four time points. Of these, DEGs involved in carbon metabolism, ion transport, and ROS metabolism and the groups which they belonged to were identified, which demonstrated their time-dependent response mechanisms. Overall, the transcriptome analysis shed light on the salinity stress response mechanisms in A. officinalis and provided a basis for future studies on salt-tolerance molecular improvement.










Similar content being viewed by others
References
Almeida DM, Margarida Oliveira M, Saibo NJM (2017) Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet Mol Biol 40:326–345
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Ashraf M, Harris PJC (2004) Potential biochemical indicators of salinity tolerance in plants. Plant Sci 166:3–16
Assaha DVM, Ueda A, Saneoka H, Al-Yahyai R, Yaish MW (2017) The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol 8:509
Bernardi P, Lippe G (2019) Editorial: structure and function of F- and V-ATPases. Front Physiol 10:358
Bose J, Munns R, Shabala S, Gilliham M, Pogson B, Tyerman SD (2017) Chloroplast function and ion regulation in plants growing on saline soils: lessons from halophytes. J Exp Bot 68:3129–3143
Cao YP, Dai P, Dai SY (2014) Effects of salt stress on the growth of Asparagus officinalis L. seedlings and on Na+, K+ and Ca2+ distribution in them. J Southwest Univ (Nat Sci Ed) 36:31–36 (in Chinese)
Cao YP, Dai P, Dai SY (2017) Effects of arbuscular mycorrhiza fungi (AMF) on osmoregulation substances and antioxidant enzyme activities of asparagus plant under salt stress. J Southwest Univ (Nat Sci Ed) 39:43–48 (in Chinese)
Chaves MM, Flexas J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 103:551–560
Dong Y, Fan G, Zhao Z, Xu E, Deng M, Wang L, Niu S (2017) Transcriptome-wide profiling and expression analysis of two accessions of Paulownia australis under salt stress. Tree Genet Genomes 13:97
ElSayed AI, Rafudeen MS, Golldack D (2014) Physiological aspects of raffinose family oligosaccharides in plants: protection against abiotic stress. Plant Biol 16:1–8
Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu MC, Maman J, Steinhorst L, Schmitz-Thom I, Yvon R, Kudla J, Wu HM, Cheung AY, Dinneny JR (2018) The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol 28:666–675
Fernie AR, Carrari F, Sweetlove LJ (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7:254–261
Flowers TJ, Munns R, Colmer TD (2015) Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann Bot 115:419–431
Foyer CH, Noctor G (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155:2–18
George S, Manoharan D, Li J, Britton M, Parida A (2018) Drought and salt stress in Macrotyloma uniflorum leads to common and specific transcriptomic responses and reveals importance of raffinose family oligosaccharides in stress tolerance. Gene Rep 10:7–16
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930
Gruber MY, Xia J, Yu M, Steppuhn H, Wall K, Messer D, Sharpe AG, Acharya SN, Wishart DS, Johnson D, Miller DR, Taheri A (2017) Transcript analysis in two alfalfa salt tolerance selected breeding populations relative to a non-tolerant population. Genome 60:104–127
Gu C, Xu S, Wang Z, Liu L, Zhang Y, Deng Y, Huang S (2018) De novo sequencing, assembly, and analysis of Iris lactea var. chinensis roots’ transcriptome in response to salt stress. Plant Physiol Biochem 125:1–12
Ji X, Van den Ende W, Van Laere A, Cheng S, Bennett J (2005) Structure, evolution, and expression of the two invertase gene families of rice. J Mol Evol 60:615–634
Jiang Z, Zhou X, Tao M, Yuan F, Liu L, Wu F, Wu X, Xiang Y, Niu Y, Liu F, Li C, Ye R, Byeon B, Xue Y, Zhao H, Wang HN, Crawford BM, Johnson DM, Hu C, Pei C, Zhou W, Swift GB, Zhang H, Vo-Dinh T, Hu Z, Siedow JN, Pei ZM (2019) Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572:341–346
Karami A, Sepehri A (2018) Beneficial role of MWCNTs and SNP on growth, physiological and photosynthesis performance of barley under NaCl stress. J Soil Sci Plant Nutr 18:752–771
Kito K, Yamane K, Yamamori T, Matsuhira H, Tanaka Y, Takabe T (2018) Isolation, functional characterization and stress responses of raffinose synthase genes in sugar beet. J Plant Biochem Biotechnol 27:36–45
Kong F, Li H, Sun P, Zhou Y, Mao Y (2014) De novo assembly and characterization of the transcriptome of seagrass Zostera marina using Illumina paired-end sequencing. PLoS One 9:e112245
Liu A, Xiao Z, Li MW, Wong FL, Yung WS, Ku YS, Wang Q, Wang X, Xie M, Yim AKY, Chan TF, Lam HM (2018) Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ 42:98–114
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25:402–408
Mahajan MM, Goyal E, Singh AK, Gaikwad K, Kanika K (2017) Transcriptome dynamics provide insights into long-term salinity stress tolerance in Triticum aestivum cv. Kharchia Local. Plant Physiol Biochem 121:128–139
Manishankar P, Wang N, Köster P, Alatar AA, Kudla J (2018) Calcium signaling during salt stress and in the regulation of ion homeostasis. J Exp Bot 69:4215–4226
Mansour MMF (2014) The plasma membrane transport systems and adaptation to salinity. J Plant Physiol 171:1787–1800
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33:453–467
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Munns R, Gilliham M (2015) Salinity tolerance of crops—what is the cost? New Phytol 208:668–673
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Nakamura A, Fukuda A, Sakai S, Tanaka Y (2006) Molecular cloning, functional expression and subcellular localization of two putative vacuolar voltage-gated chloride channels in rice (Oryza sativa L.). Plant Cell Physiol 47:32–42
Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147:1251–1263
Obata T, Fernie AR (2012) The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci 69:3225–3243
Parihar P, Singh S, Singh R, Singh VP, Prasad SM (2015) Effect of salinity stress on plants and its tolerance strategies: a review. Environ Sci Pollut Res 22:4056–4075
Peterbauer T, Lahuta LB, Blöchl A, Mucha J, Jones DA, Hedley CL, Gòrecki RJ, Richter A (2001) Analysis of the raffinose family oligosaccharide pathway in pea seeds with contrasting carbohydrate composition. Plant Physiol 127:1764–1772
Plaxton WC (1996) The organization and regulation of plant glycolysis. Annu Rev Plant Physiol Plant Mol Biol 47:185–214
Qiu J, Henderson SW, Tester M, Roy SJ, Gilliham M (2016) SLAH1, a homologue of the slow type anion channel SLAC1, modulates shoot Cl- accumulation and salt tolerance in Arabidopsis thaliana. J Exp Bot 67:4495–4505
Rahman H, Jagadeeshselvam N, Valarmathi R, Sachin B, Sasikala R, Senthil N, Sudhakar D, Robin S, Muthurajan R (2014) Transcriptome analysis of salinity responsiveness in contrasting genotypes of finger millet (Eleusine coracana L.) through RNA-sequencing. Plant Mol Biol 85:485–503
Roy SJ, Negrão S, Tester M (2014) Salt resistant crop plants. Curr Opin Biotechnol 26:115–124
Sairam RK, Tyagi A (2004) Physiology and molecular biology of salinity stress tolerance in plants. Curr Sci 86:407–421
Shao F, Zhang L, Wilson IW, Qiu D (2018) Transcriptomic analysis of Betula halophila in response to salt stress. Int J Mol Sci 19:3412
Skorupa M, Gołębiewski M, Domagalski K, Kurnik K, Abu Nahia K, Złoch M, Tretyn A, Tyburski J (2016) Transcriptomic profiling of the salt stress response in excised leaves of the halophyte Beta vulgaris ssp. maritime. Plant Sci 243:56–70
Sui N, Yang Z, Liu M, Wang B (2015) Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genomics 16:534
Sui N, Wang Y, Liu S, Yang Z, Wang F, Wan S (2018) Transcriptomic and physiological evidence for the relationship between unsaturated fatty acid and salt stress in peanut. Front Plant Sci 9:7
Sun Z, Qi X, Wang Z, Li P, Wu C, Zhang H, Zhao Y (2013) Overexpression of TsGOLS2, a galactinol synthase, in Arabidopsis thaliana enhances tolerance to high salinity and osmotic stresses. Plant Physiol Biochem 69:82–89
Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29:417–426
Thalmann M, Santelia D (2017) Starch as a determinant of plant fitness under abiotic stress. New Phytol 214:943–951
Van Zelm E, Zhang Y, Testerink C (2020) Salt tolerance mechanisms of plants. Annu Rev Plant Biol 71:24.1–24.31
Verma AK, Upadhyay SK, Verma PC, Solomon S, Singh SB (2011) Functional analysis of sucrose phosphate synthase (SPS) and sucrose synthase (SS) in sugarcane (Saccharum) cultivars. Plant Biol 13:325–332
Wang Z, Liu Y, Wei J, Deng X (2012) Cloning and expression of a gene encoding a raffinose synthase in the resurrection plant Boea hygrometrica. Chin Bull Bot 47:44–54 (in Chinese)
Wang N, Qian Z, Luo M, Fan S, Zhang X, Zhang L (2018) Identification of salt stress responding genes using transcriptome analysis in green alga Chlamydomonas reinhardtii. Int J Mol Sci 19:3359
Wang Q, Cao K, Zhu G, Fang W, Chen C, Wang X, Wang L (2019) Comparative transcriptome analysis of genes involved in the response of resistant and susceptible peach cultivars to water stress. Sci Hortic 245:29–38
Yang Y, Guo Y (2018) Unraveling salt stress signaling in plants. J Integr Plant Biol 60:796–804
Yang H, Sun M, Lin S, Guo Y, Yang Y, Zhang T, Zhang J (2017) Transcriptome analysis of Crossostephium chinensis provides insight into the molecular basis of salinity stress responses. PLoS One 12:e0187124
Yang Z, Li JL, Liu LN, Xie Q, Sui N (2020) Photosynthetic regulation under salt stress and
Yuan F, Leng B, Wang B (2016) Progress in studying salt secretion from the salt glands in recretohalophytes: how do plants secrete salt? Front Plant Sci 7:997
Yue C, Cao H, Wang L, Zhou Y, Hao X, Zeng J, Wang X, Yang Y (2014) Molecular cloning and expression analysis of tea plant aquaporin (AQP) gene family. Plant Physiol Biochem 83:65–76
Zhang H, Han B, Wang T, Chen S, Li H, Zhang Y, Dai S (2012) Mechanisms of plant salt response: insights from proteomics. J Proteome Res 11:49–67
Zhang Q, Song X, Bartels D (2018) Sugar metabolism in the desiccation tolerant grass Oropetium thomaeum in response to environmental stresses. Plant Sci 270:30–36
Zheng L, Meng Y, Ma J, Zhao X, Cheng T, Ji J, Chang E, Meng C, Deng N, Chen L, Shi S, Jiang Z (2015) Transcriptomic analysis reveals importance of ROS and phytohormones in response to short-term salinity stress in Populus tomentosa. Front Plant Sci 6:678
Zhong M, Yuan Y, Shu S, Sun J, Guo S, Yuan R, Tang Y (2015) Effects of exogenous putrescine on glycolysis and Krebs cycle metabolism in cucumber leaves subjected to salt stress. Plant Growth Regul 79:319–330
Zhou J, Wang X, Jiao Y, Qin Y, Liu X, He K, Chen C, Ma L, Wang J, Xiong L, Zhang Q, Fan L, Deng XW (2007) Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol 63:591–608
Zhou ML, Zhang Q, Zhou M, Sun ZM, Zhu XM, Shao JR, Tang YX, Wu YM (2012) Genome-wide identification of genes involved in raffinose metabolism in maize. Glycobiology 22:1775–1785
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zhu H, Yu X, Xu T, Wang T, Du L, Ren G, Dong K (2015) Transcriptome profiling of cold acclimation in bermudagrass (Cynodon dactylon). Sci Hortic 194:230–236
Funding
This work was supported by the National Natural Science Foundation of China (No. 31960314), Key R&D Projects of Hebei Province (No. 19226334D), Applied Basic Research Projects of Yunnan Province (No. 2018FG001-028), Scientific and Technological Innovation Projects of Hebei Province (No. 20193-01-01), and “Giant Project” of Hebei Province (No. 2018-3).
Author information
Authors and Affiliations
Contributions
Yanpo Cao designed the project and performed the experiments. Xuhong Zhang and Changzhi Han analyzed the data and wrote the manuscript. All authors contributed to the revision of this manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
• RNA-Seq analysis of asparagus leaves under salt stress was performed.
• Total 4803 DEGs and their temporal expression patterns were identified.
• DEGs related to carbon metabolism, ion transport, and ROS metabolism may be crucial.
Rights and permissions
About this article
Cite this article
Zhang, X., Han, C. & Cao, Y. Transcriptomic and Physiological Analyses Reveal the Dynamic Response to Salinity Stress of the Garden Asparagus (Asparagus officinalis L.). Plant Mol Biol Rep 38, 613–627 (2020). https://doi.org/10.1007/s11105-020-01226-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01226-x