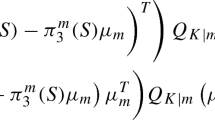Abstract
A fast Bayesian method that seamlessly fuses classification and hypothesis testing via discriminant analysis is developed. Building upon the original discriminant analysis classifier, modelling components are added to identify discriminative variables. A combination of cake priors and a novel form of variational Bayes we call reverse collapsed variational Bayes gives rise to variable selection that can be directly posed as a multiple hypothesis testing approach using likelihood ratio statistics. Some theoretical arguments are presented showing that Chernoff-consistency (asymptotically zero type I and type II error) is maintained across all hypotheses. We apply our method on some publicly available genomics datasets and show that our method performs well in practice for its computational cost. An R package VaDA has also been made available on Github.




Similar content being viewed by others
References
Ahdesmäki, M., Strimmer, K.: Feature selection in omics prediction problems using CAT score and false discovery rate control. Ann. Appl. Stat. 4(1), 503–519 (2010)
Alizadeh, A., Eisen, M., Davis, R., Ma, C., Lossos, I., Rosenwald, A., Boldrick, J., Sabet, H., Tran, T., Yu, X., Powell, J., Yang, L., Marti, G., Moore, T., Hudson, J.J., Lu, L., Lewis, D., Tibshirani, R., Sherlock, G., Chan, W., Greiner, T., Weisenburger, D., Armitage, J., Warnke, R., Levy, R., Wilson, W., Grever, M., Byrd, J., Botstein, D., Brown, P., Staudt, L.: Distinct types of diffuse large b-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000)
Alon, U., Barkai, N., Notterman, D., Gish, K., Ybarra, S., Mack, D., AJ, L.: Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. 96(12), 6745–6750 (1999)
Benjamini, Y., Daniel, Y.: The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29(4), 1165–1188 (2001)
Benjamini, Y., Hochberg, Y.: Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57(1), 289–300 (1995)
Bickel, P.J., Levina, E.: Some theory for Fisher’s linear discriminant function, ‘Naïve Bayes’ and some alternatives when there are many more variables than observations. Bernoulli 10(6), 989–1010 (2004)
Blei, D.M., Jordan, M.I.: Variational inference for Dirichlet processes. Bayesian Anal. 1(1), 121–144 (2006)
Blei, D.M., Kucukelbir, A., McAuliffe, J.D.: Variational inference: a review for statisticians. J. Am. Stat. Assoc. 112(518), 859–877 (2017)
Bonferroni, C.E.: Teoria statistica delle classi e calcolo delle probabilità. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze (1936)
Breiman, L.: Random forests. Mach. Learn. 45(1), 5–32 (2001)
Cai, T., Liu, W.: A direct estimation approach to sparse linear discriminant analysis. J. Am. Stat. Assoc. 106(496), 1566–1577 (2011)
Carvalho, C.M., Polson, N.G., Scott, J.G.: The horseshoe estimator for sparse signals. Biometrika 97(2), 465–480 (2010)
Chen, Y., Feng, J.: Efficient method for Moore–Penrose inverse problems involving symmetric structure based on group theory. J. Comput. Civ. Eng. 28(2), 182–190 (2014)
Chicco, D.: Ten quick tips for machine learning in computational biology. BioData Min. 10(35), 1–17 (2017)
Clemmensen, L.: On Discriminant Analysis Techniques and Correlation Structures in High Dimensions. Technical report-2013(4). Technical University of Denmark (DTU), Kgs. Lyngby (2013)
Clemmensen, L., Kuhn, M.: sparseLDA: Sparse Discriminant Analysis. R package version 0.1-9 (2016)
Clemmensen, L., Witten, D., Hastie, T., Ersboll, B.: Sparse discriminant analysis. Technometrics 53(4), 406–413 (2011)
Cortes, C., Vapnik, V.: Support-vector networks. Mach. Learn. 20(3), 273–297 (1995)
Courrieu, P.: Fast computation of Moore–Penrose inverse matrices. Neural Inf. Process. Lett. Rev. 8(2), 25–29 (2005)
Craig-Shapiro, R., Kuhn, M., Xiong, C., Pickering, E.H., Liu, J., Misko, T.P., Perrin, R.J., Bales, K.R., Soares, H., Fagan, A.M., David, M.H.: Multiplexed immunoassay panel identifies novel CSF biomarkers for Alzheimer’s disease diagnosis and prognosis. PLoS ONE 6, e18850 (2011)
Donoho, D., Jin, J.: Higher criticism for detecting sparse heterogeneous mixtures. Ann. Stat. 32(3), 962–994 (2004)
Donoho, D., Jin, J.: Higher criticism thresholding. optimal feature selection when useful features are rare and weak. Proc. Natl. Acad. Sci. 105(39), 14790–14795 (2008)
Duarte Silva, P.A.: Two group classification with high-dimensional correlated data: a factor model approach. Comput. Stat. Data Anal. 55(11), 2975–2990 (2011)
Duarte Silva, P.A.: HiDimDA: High Dimensional Discriminant Analysis. R package version 0.2-4 (2015)
Dudoit, S., Fridyland, J., Speed, T.P.: Comparison of discrimination methods for classification of tumours using gene expression data. J. Am. Stat. Assoc. 97(457), 77–87 (2002)
Eddelbuettel, D.: Seamless R and C++ Integration with Rcpp. Springer, Berlin (2013)
Erickson, B.J., Kirk, S., Lee, Y., Bathe, O., Kearns, M., Gerdes, C., Rieger-Christ, K., Lemmerman, J.: Radiology data from The Cancer Genome Atlas Liver Hepatocellular Carcinoma [TCGA-LIHC] collection. The Cancer Imaging Archive (2016). http://doi.org/10.7937/K9/TCIA.2016.IMMQW8UQ
Fan, J., Fan, Y.: High-dimensional classification using features annealed independence rules. Ann. Stat. 36(6), 2605–2637 (2008)
Fan, J., Lv, J.: A selective overview of variable selection in high dimensional feature space. Stat. Sin. 20(1), 101–148 (2010)
Fernández-Delgado, M., Cernadas, E., Barro, S.: Do we need hundreds of classifiers to solve real world classification problems? J. Mach. Learn. 15, 3133–3181 (2014)
Fisher, R.A.: The use of multiple measurements in taxonomic problems. Ann. Eugen. 7(2), 179–188 (1936)
Fisher, T., Sun, X.: Improved stein-type shrinkage estimators for the high-dimensional multivariate normal covariance matrix. Comput. Stat. Data Anal. 55(1), 1909–1918 (2011)
Friedman, J.H.: Regularized discriminant analysis. J. Am. Stat. Assoc. 84(405), 165–175 (1989)
Friguet, C., Kloareg, M., Causeur, D.: A factor model approach to multiple testing under dependence. J. Am. Stat. Assoc. 104(488), 1406–1415 (2009)
Genuer, R., Poggi, J., Tuleau-Malot, C., Villa-Vialaneix, N.: Random forests for big data. Big Data Res. 9, 28–46 (2017)
Golub, T., Slonim, D., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J.: Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286(5439), 531–537 (1999)
Guo, Y., Hastie, T., Tibshirani, R.: Regularized linear discriminant analysis and its application in microarrays. Biostatistics 8(1), 86–100 (2007)
Guo, Y., Hastie, T., Tibshirani, R.: RDA: Shrunken Centroids Regularized Discriminant Analysis. R package version 1.0.2-2.1 (2018)
Hastie, T., Tibshirani, R., Narasimhan, B., Chu, G.: PAMR: Prediction Analysis for Microarrays. R package version 1.55 (2014)
Helleputte, T.: LiblineaR: Linear Predictive Models Based on the LIBLINEAR C/C++ Library. R package version 2.10-8 (2017)
Jordan, M.I.: On statistics, computation and scalability. Bernoulli 19(4), 1378–1390 (2013)
Jorissen, R.N., Lipton, L., Gibbs, P., Chapman, M., Desai, J., Jones, I.T., Yeatman, T.J., East, P., Tomlinson, I.P., Verspaget, H.W., Aaltonen, L.A., Kruhoffer, M., Orntoft, T.F., Andersen, C.L., Sieber, O.M.: DNA copy-number alterations underlie gene expression differences between microsatellite stable and unstable colorectal cancers. Clin. Cancer Res. 14(24), 8061–8069 (2008)
Kuhn, M., Wing, J., Weston, S., Williams, A., Keefer, C., Engelhardt, A., Cooper, T., Mayer, Z., Kenkel, B., Benesty, M., Lescarbeau, R., Ziem, A., Scrucca, L., Tang, Y., Candan, C., Hunt, T.: Caret: Classification and Regression Training. R package version 6.0-84 (2019)
Lim, T.S., Loh, W.Y., Shih, Y.S.: A comparison of prediction accuracy, complexity, and training time of thirty-three old and new classification algorithms. Mach. Learn. 40, 203–229 (2000)
Liu, J.J., Cutler, G., Li, W., Pan, Z., Peng, S., Hoey, T., Chen, L., Ling, X.B.: Multiclass cancer classification and biomarker discovery using GA-based algorithms. Bioinformatics 21(11), 2691–2697 (2005)
Luts, J., Ormerod, J.T.: Mean field variational Bayesian inference for support vector machine classification. Comput. Stat. Data Anal. 73, 163–176 (2014)
Mai, Q., Zou, H., Yuan, M.: A direct approach to sparse discriminant analysis in ultra-high dimensions. Biometrika 99(1), 29–42 (2012)
Mai, Q., Yang, Y., Zou, H.: Multiclass sparse discriminant analysis. arXiv (2015)
Marks, S., Dunn, O.: Discriminant functions when covariance matrices are unequal. J. Am. Stat. Assoc. 69(346), 555–559 (1974)
Matthews, B.W.: Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta (BBA) Protein Struct. 405(2), 442–451 (1975)
Ormerod, J.T., Wand, M.P.: Explaining variational approximations. Am. Stat. 64(2), 140–153 (2010)
Ormerod, J.T., Stewart, M., Yu, W., Romanes, S.: Bayesian hypothesis test with diffused priors: Can we have our cake and eat it too? arXiv (2017)
Pan, R., Wang, H.O., Li, R.: Ultrahigh-dimensional multiclass linear discriminant analysis by pairwise sure independence screening. J. Am. Stat. Assoc. 111(513), 169–179 (2016)
Perthame, E., Friguet, C., Causeur, D.: Stability of feature selection in classification issues for high-dimensional correlated data. Stat. Comput. 26(4), 783–796 (2016)
Perthame, E., Friguet, C., Causeur, D.: FADA: Variable Selection for Supervised Classification in High Dimension. R package version 1.3.3 (2018)
Reif, M., Shafait, F., Dengel, A.: Prediction of classifier training time including parameter optimization. Annu. Conf. Artif. Intell. KI2011, 260–271 (2011)
Safo, S.E., Ahn, J.: General sparse multi-class linear discriminant analysis. Comput. Stat. Data Anal. 99, 81–90 (2016)
Shaffer, J.P.: Multiple hypothesis testing. Annu. Rev. Psychol. 46, 561–584 (1995)
Shao, J., Wang, Y., Deng, X., Wang, S.: Sparse linear discriminant analysis with applications to high dimensional data. Ann. Stat. 39(2), 1241–1265 (2011)
Singh, D., Febbo, P., Ross, K., Jackson, D., Manola, J., Ladd, C., Tamayo, P., Renshaw, A., DAmico, A., Richie, J., Lander, E., Loda, M., Kantoff, P., Golub, T.: Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1(2), 203–209 (2002)
Srivastava, S., Gupta, M.R., Frigyik, B.A.: Bayesian quadratic discriminant analysis. J. Mach. Learn. Res. 8(Jun), 1277–1305 (2007)
Storey, J.D.: The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 31(6), 2013–2035 (2003)
Teh, Y.W., Newman, D., Welling, M.: A collapsed variational Bayesian inference algorithm for latent Dirichlet allocation. In: Jordan, M.I., LeCun, Y., Solla, S.A. (eds.) Advances in Neural Information Processing Systems, vol. 19, pp. 1353–1360. MIT Press, Cambridge (2007)
Thomaz, C., Kitani, E., Gillies, D.: A maximum uncertainty lda-based approach for limited sample size problems—with applications to face recognition. J. Braz. Comput. Soc. 12(2), 7–18 (2006)
Tibshirani, R., Hastie, T., Narasimhan, B., Chu, G.: Class prediction by nearest shrunken centroids, with applications to DNA microarrays. Stat. Sci. 18(1), 104–117 (2003)
van der Maaten, L., Hinton, G.: Visualising data using t-sne. J. Mach. Learn. Res. 9, 2579–2605 (2017)
Wang, Y., Blei, D.: Frequentist consistency of variational bayes. J. Am. Stat. Assoc. 9, 1–15 (2018). https://doi.org/10.1080/01621459.2018.1473776
Witten, D.: Classification and clustering of sequencing data using a Poisson model. Ann. Appl. Stat. 5(4), 2493–2518 (2011)
Witten, D.: penalizedLDA: Penalized Classification Using Fisher’s Linear Discriminant. R package version 1.1 (2015)
Witten, D., Tibshirani, R.: Penalized classification using Fisher’s linear discriminant. J. R. Stat. Soc. Ser. B 73(5), 754–772 (2011)
Xu, P., Brock, G.N., Parrish, R.S.: Modified linear discriminant analysis approaches for classification of high-dimensional microarray data. Comput. Stat. Data Anal. 53, 1674–1687 (2009)
Zavorka, S., Perrett, J.: Minimum sample size considerations for two-group linear and quadratic discriminant analysis with rare populations. Commun. Stat. Simul. Comput. 43(7), 1726–1739 (2014)
Zhang, A., Zhou, H.: Theoretical and computational guarantees of mean field variational inference for community detection. arXiv (2017)
Zhang, C., Liu, C., Zhang, X., Almpanidis, G.: An up-to-date comparison of state-of-the-art classification algorithms. Exp. Syst. Appl. 82, 128–150 (2017)
Acknowledgements
The authors would like to thank Rachel Wang (University of Sydney), the associate editor, and the anonymous reviewers for their valuable feedback to greatly improve this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix
Appendix
1.1 VQDA derivations
In the VQDA setting (\(\sigma _{j 1}^2 \ne \sigma _{j 0}^2\)) the posterior distribution of \(\gamma _j\) given \({\mathcal {D}}\) and \(\mathbf{x}_{n+1}\) may be expressed as
By letting \(h \rightarrow \infty \), the marginal likelihood of the data in the denominator is of the same form as Eq. (9) with the exception that
and
where
and the \(j{\text {th}}\) entry of \({\varvec{\lambda }}_{\text {Bayes}}\) is (as \(h \rightarrow \infty \))
where \(\xi (x) = \log \varGamma (x) + x - x \log (x) - \tfrac{1}{2} \log (2 \pi )\). Since the calculation of the marginal likelihood involves a combinatorial sum over \(2^{p+1}\) binary combinations, exact Bayesian inference is also computationally impractical in the VQDA setting.
Similar to VLDA, we will use RCVB to approximate the posterior \(p({\varvec{\gamma }}, y_{n+1} | \mathbf{x}, \mathbf{x}_{n+1}, \mathbf{y})\) by
This yields the approximate posterior for \(\gamma _j\) as
For a sufficiently large n, we can avoid the need to evaluate the expectation \( {\mathbb {E}}_{-q_j} \Big \{ {\varvec{\lambda }}_{\text {Bayes}} (\widetilde{\mathbf{x}}_j, x_{n+1,j}, \mathbf{y}, y_{n+1}) \Big \}\) by applying Taylor’s expansion to obtain the approximation
and, similar to VLDA, \({\varvec{\lambda }}_{\text {Bayes}}\) does not depend on the new observation \((\mathbf{x}_{n+1}, y_{n+1})\). By using the approximation in (16), we have
To obtain the approximate density for \(y_{n+1}\), we integrate analytically over \({\varvec{\theta }}_1\) to obtain
where the \(j{\text {th}}\) element of the \(p \times 1\) vector \({\varvec{\phi }}(\mathbf{x}_{n+1}; \widehat{{\varvec{\mu }}}_k, \widehat{{\varvec{\sigma }}}_{k}^2)\) is the Gaussian density
and the \(\log \) prefix denotes an element-wise \(\log \) of a vector.
In the general case with m new observations, we may apply Taylor’s expansion results from (16) to compute the approximate classification probability for \(y_{n+i}\) as
The RCVB algorithm for VQDA may be found in Table 2.
Rights and permissions
About this article
Cite this article
Yu, W., Ormerod, J.T. & Stewart, M. Variational discriminant analysis with variable selection. Stat Comput 30, 933–951 (2020). https://doi.org/10.1007/s11222-020-09928-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11222-020-09928-8