Abstract
H1N1, a major pathogenic subtype of influenza A virus, causes a respiratory infection in humans and livestock that can range from a mild infection to more severe pneumonia associated with acute respiratory distress syndrome. Understanding the dynamic changes in the genome and the related functional changes induced by H1N1 influenza virus infection is essential to elucidating the pathogenesis of this virus and thereby determining strategies to prevent future outbreaks. In this study, we filtered the significantly expressed genes in mouse pneumonia using mRNA microarray analysis. Using STC analysis, seven significant gene clusters were revealed, and using STC-GO analysis, we explored the significant functions of these seven gene clusters. The results revealed GOs related to H1N1 virus-induced inflammatory and immune functions, including innate immune response, inflammatory response, specific immune response, and cellular response to interferon-beta. Furthermore, the dynamic regulation relationships of the key genes in mouse pneumonia were revealed by dynamic gene network analysis, and the most important genes were filtered, including Dhx58, Cxcl10, Cxcl11, Zbp1, Ifit1, Ifih1, Trim25, Mx2, Oas2, Cd274, Irgm1, and Irf7. These results suggested that during mouse pneumonia, changes in the expression of gene clusters and the complex interactions among genes lead to significant changes in function. Dynamic gene expression analysis revealed key genes that performed important functions. These results are a prelude to advancements in mouse H1N1 influenza virus infection biology, as well as the use of mice as a model organism for human H1N1 influenza virus infection studies.
Similar content being viewed by others
Introduction
Influenza A virus is a single negative-stranded RNA virus belonging to the Orthomyxoviridae family that can cause localized outbreaks of infection or worldwide epidemics. It is characterized by its high infectivity, rapid spread and wide host range. Even in non-pandemic years, influenza A viruses infect 5–15% of the global population and result in >500,000 deaths per year [1]. Influenza A virus infection is a major cause of disease-related mortality. According to the antigenicity of surface structural proteins (hemagglutinin and neuraminidase), influenza A viruses are divided into different subtypes (such as H1N1, H7N9, H5N1). Not all H1N1 are highly pathogenic and indeed pathogenicity is host dependent. H1N1 influenza virus infects both humans and livestock causing respiratory disease ranging from mild infections to more severe pneumonia associated with acute respiratory distress syndrome. In 2009, human infection with a novel H1N1 influenza virus of swine origin was reported and rapidly spread worldwide, leading the World Health Organization (WHO) to declare on 11 June 2009 the first influenza pandemic in more than 40 years [2]. Understanding the dynamic changes in the expression of genes and the corresponding functional changes induced by H1N1 influenza virus infection is essential to understanding the pathogenesis of this virus and thereby helping to predict and control future outbreaks.
In this study, we developed a mouse pneumonia model of H1N1 influenza virus infection. To study the dynamic changes in gene expression during the process of lesion development, we inspected two time points (days 2 and 5) of H1N1 influenza virus infection. The series test of cluster (STC) algorithm of gene expression dynamics was used to profile the gene expression time series and to identify the seven most probable set of clusters generating the observed time series. STC-gene ontology (GO) analysis was used to analyze the main functions of the seven clusters, and one cluster was identified to play a key role in the mouse pneumonia model. The dynamic regulation relationship of the key genes in mouse pneumonia was investigated by dynamic-gene network analysis, and the most important genes were filtered, including Dhx58, Cxcl10, Cxcl11, Zbp1, Ifit1, Ifih1, Trim25, Mx2, Oas2, Cd274, Irgm1, and Irf7. It is worth noting that Cxcl10 and Cxcl11 were chemokine ligands; Ifit1, Ifih1 and Irf7 were related to interferon function; and Mx2 was a natural barrier for influenza virus replication in the host. These findings lay the foundation for future in-depth analyses.
Materials and methods
Virus
The virus strain used in this study was A/Puerto Rico/8/34 (PR8, H1N1) (referred to as PR8 virus below, ATCC, Virginia, USA), a well-characterized mouse-adapted laboratory strain of influenza A virus used as the genetic backbone for viruses from which inactivated influenza virus vaccines are generated. The virus was grown in the allantoic cavities of 10-day-old embryonated chicken eggs. Allantoic fluid containing virus was harvested and stored in aliquots at −80 °C until use. The 50% tissue culture infectious dose (TCID50) was determined by serial dilution of the virus in Madin–Darby canine kidney (MDCK) cells (ATCC, Manassas, VA, USA) and calculated by the method developed by Reed and Muench [3]. All experiments with live influenza viruses were performed in an approved biosafety level-2 laboratory.
Mouse pneumonia model of PR8 virus infection
ICR mice have characteristics with adaptability, physical robustness, strong fecundity, fast growth, good reproducibility of the experiment, and are more commonly used in replicating pathological models. ICR mice have susceptibility to influenza viruses. Specific-pathogen-free 13–15 g male and female ICR mice (equal numbers) were provided by Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). PR8 virus infections in mice were conducted as described previously [4]. Briefly, the mice (n = 12/group) were anesthetized with ether and were intranasally inoculated with 104 TCID50 of PR8 virus in physiological saline. Based on the pilot study, weight loss and lung damage were observed in mice infected with 104 TCID50 of PR8 virus. In addition, a normal control group (n = 12) was given intranasal physiological saline. Mice were sacrificed after weighing; entire lungs from randomly selected mice infected with PR8 virus (n = 12/group) were harvested on days 2 and 5; and entire lungs from 12 randomly selected control mice were also harvested on day 5. The lung index was used to evaluate the degree of damage of the mouse lungs by PR8 virus. The lung index of mice was calculated using the formula: lung index (%) = (lung weight/body weight) × 100%.
RNA isolation
For total RNA extraction, whole mouse lung tissues were homogenized in liquid nitrogen. Total RNAs were extracted from mouse lungs using the Qiagen miRNeasy Mini Kit (Qiagen, Hilden, Germany).
MRNA microarray analysis
Total RNAs were processed by mRNA microarray analysis using an Affymetrix GeneChip Mouse 1.0 (Affymetrix, Santa Clara, CA, USA). For each group, three biological replicates were performed, and each replicate sample came from four mice. The random variance model (RVM) and F-test were applied to filter the differentially expressed genes for the control and temporal groups, because the RVM F-test can raise degrees of freedom effectively in the case of small samples. After significance analysis and false discovery rate (FDR) analysis, the differentially expressed genes were selected according to their p value threshold [5–7].
STC analysis
The STC algorithm of gene expression dynamics was used to profile the gene expression time series and to identify the most probable set of clusters generating the observed time series. This method explicitly took into account the dynamic nature of the temporal gene expression profiles during clustering and identified the number of distinct clusters.
We selected differentially expressed genes at a logical sequence according to RVM corrective analysis of variance (ANOVA). In accordance with the different signal density change tendencies of genes under different situations, we identified a set of unique model expression tendencies. The raw expression values were converted into log2 ratios. Using a strategy for clustering time-series gene expression data, we defined some unique profiles. The expression model profiles are related to the actual or the expected number of genes assigned to each model profile. Significant profiles have higher probabilities than expected by the Fisher’s exact test and multiple comparison tests [8–10].
STC-GO analysis
GO analysis was applied to analyze the main function of the differentially expressed genes according to their GO, which is the key functional classification system employed by NCBI, which can organize genes into hierarchical categories and uncover the gene regulatory networks on the basis of biological processes and molecular functions [11, 12].
GO analysis is applied to the genes showing certain specific tendencies. It is used to find the main function of the genes that display the same expression tendencies according to NCBI GO. Generally, Fisher’s exact test and χ 2 test were used to classify the GO category, and the FDR was calculated to correct the p-value [13]; the smaller the FDR, the smaller the error in judging the p-value. The FDR was defined as FDR = 1−N k/T, where N k refers to the number of Fisher’s test p-values less the χ 2 test p-values. We computed p-values for the GOs of all the differentially expressed genes. Enrichment provides a measure of the significance of the function: as the enrichment increases, the corresponding function is more specific, which helps to identify those GOs with a more definite functional description in the experiment. Within the significant category, the enrichment R e was given by R e = (n f /n)/(N f/N), where n f is the number of differential genes within the particular category, n is the total number of genes within the same category, N f is the number of differential genes in the entire microarray, and N is the total number of genes in the microarray [14].
Pathway analysis and STC-pathway analysis
Pathway analysis was used to determine the significant pathway of the differential genes according to KEGG, Biocarta and Reatome. We used Fisher’s exact test and χ 2 test to select the significant pathway, and the threshold of significance was defined by the p-value and the FDR. The enrichment Re was calculated using the equation detailed above [15–17].
According to the result of STC analysis, STC-pathway analysis was used to determine the significant pathway of the genes that show the same expression trend according to KEGG, Biocarta, and Reatome. We used the Fisher’s exact test and the χ2 test to select the significant pathway, and the threshold of significance was defined by the p-value and the FDR. The enrichment R e was calculated as described above [8, 9, 18, 19].
Dynamic-gene network
Within the network analysis, nodes represent the genes, and the edges between genes depict the interactions between them [20]. All the nodes were marked with degrees, which were defined as the link numbers that one node has to the other. Genes with higher degrees occupied more central positions in the network and had a stronger capacity for modulating adjacent genes. In addition, k-core in graph theory was applied to describe the characteristics of the network including, but not limited to, the centrality of genes within a network and the complexity of the subnetworks. According to the relationship between genes, they were divided into several subnetworks, and were marked with different colors [14].
A dynamic-gene network was built according to the normalized signal intensity of specific expression genes. At first, the correlation of each pair of genes was calculated as the basis for choosing the significant correlation of gene pairs. Then the gene–gene interaction network was established according to the correlation between genes. Within the network, nodes represent the genes and the edges between genes depict the interactions between them. All the nodes were marked with degrees; the degree centrality is one of the simplest and most important measures of the centrality of a gene and its relative importance within a network. Degree centrality is defined as the number of links that one node has to all the other nodes [21]. Moreover, in studying some properties of the networks, k-cores in graph theory were introduced as a method of simplifying the analysis of graph topologies. A k-core of a network is a sub-network in which all nodes are connected to at least k other genes in the sub-network. A k-core of a gene–gene interaction network usually contains cohesive groups of genes. The purpose of network structure analysis is to locate core regulatory factors (genes), in one network. Core regulatory factors connect the most adjacent genes and have the largest degrees. When considering different networks, the core regulatory factors were determined by the degree of differences between two class samples [19]. The core regulatory factors always have the largest degree differences.
Histopathologic analysis
Lungs were fixed in 4% formalin, dehydrated in ascending ethanol concentrations, embedded in paraffin, sectioned into 4-μm slices, and stained with hematoxylin and eosin (H&E). Histopathology images were recorded using a phase inverted microscope (Olympus, Tokyo, Japan).
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
qRT-PCR was performed to verify the expression levels of the Dhx58, Cxcl10, Cxcl11, Zbp1, Ifit1, Ifih1, Trim25, Mx2, Oas2, Cd274, Irgm1, and Irf7 genes in the mouse lungs. Total RNA (0.5 µg) was reverse transcribed using the Thermo Scientific RevertAid First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Rockford, IL, USA). qRT-PCR was performed on a Thermo Scientific PikoReal Real-Time PCR System (Thermo Fisher Scientific), using Thermo Scientific Maxima SYBR Green qPCR Master Mixes (Thermo Fisher Scientific). The primers used in this analysis are listed in Table S1. All samples were analyzed in triplicate, including a no-template control. The relative expression level was determined by the 2−△△Ct method, and normalized to the expression of the GAPDH gene.
Statistical analysis
The lung index results are expressed as the mean ± standard deviation (SD). Statistical analysis was performed using ANOVA for multiple comparisons, and p < 0.05 was considered statistically significant.
Results
Mouse pneumonia model of PR8 virus infection
Mice from the control, 2-day, and 5-day groups were anesthetized with ether and intranasally inoculated with 104 TCID50 PR8 virus in physiological saline. The control group was administered intranasal physiological saline. Mice were weighed and mouse lungs were harvested on days 2 and 5 after infection. Compared with the control group, lung of both infection groups lesions obviously and the lung index of both infection groups was significantly increased (p < 0.01), and when compared with the 2-day group, the lung index of the 5-day group was also significantly increased (p < 0.01) (Fig. 1a). H&E staining was performed to compare histopathological inflammatory changes. Compared with the control group, obvious inflammatory infiltration appeared in the 2- and 5-day groups (Fig. 1b). The results showed that when mice were infected with 104 TCID50 of PR8 virus, the mortality rate was zero both 2 and 5 days after infection, and weight loss and lung damage increased with time.
Mouse pneumonia model of PR8 virus infection. a Lung index of mice. Compared with the control group, the lung index of both the infection groups increased significantly (p < 0.01); and compared with the 2-day group, the lung index of the 5-day group also increased significantly (p < 0.01). b Hematoxylin and eosin staining to evaluate the histopathological inflammatory changes. Compared with the control group, significant inflammatory infiltration appeared in the 2-day and 5-day groups; arrows point to inflammatory infiltration
Differentially expressed genes in the mouse pneumonia model of PR8 virus infection as determined by microarray analysis
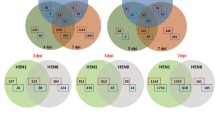
Total RNAs were extracted from the mouse lungs using the Qiagen miRNeasy Mini Kit (Qiagen). The RNA samples were prepared from four lungs from each group. mRNA expression profiles of the mouse lungs infected with PR8 virus were detected with the Affymetrix GeneChip Mouse 1.0. The RVM and F-test were applied to filter the differentially expressed genes for the control and temporal groups; after significance analysis and FDR analysis, 3371 differentially expressed genes were selected according to their p-value thresholds (p < 0.05), and the differences in mRNAs between each of the groups were displayed (Fig. 2).
mRNA microarray assay. a Cluster of control, 2-day, and 5-day group; b cluster of control and 2-day–day group; c cluster of control and 5-day group; d cluster of 2-day and 5-day group. The mRNA profiles to differentiate expression in the lungs with PR8 viral infection (2-day and 5-day) from the uninfected lungs (control). Both down-regulated (green) and up-regulated (red) mRNAs were identified in the infected lungs (Color figure online)
Genes in the mouse pneumonia model analyzed by STC
In this study, the STC algorithm of mRNA expression dynamics was used to profile 3371 differentially expressed mRNA expression time series and to identify the most probable set of clusters generating the observed time series. The 3371 mRNAs were placed into 16 possible expressive pattern profiles for STC analysis. The horizontal axis represents time points, with 2 days being located at 1.0 and 5 days being located at 2.0, and the vertical axis represents expression changes (log2 ratio). The results showed that profiles 1, 2, 3, 4, 7, 9, and 11 were significant STC (p < 0.05) (Fig. S1). In addition, mRNA expression among profiles 1, 2, and 11 was up-regulated continuously in the 5-day group and mRNA expression among profiles 4, 7, and 9 was down-regulated continuously in the 5-day group. It is worth noting that mRNA expression clusters for profiles 1 and 4 (Fig. S1, Fig. S2A), profiles 2 and 7 (Fig. S1, Fig. S2B), and profiles 11 and 9 (Fig. S1, Fig. S2C) showed the opposite trends. The difference was that mRNA expression of profile 3 was up-regulated in the 2-day group, but did not continue to be up-regulated thereafter (Fig. S1, Fig. S2D); genes of profile 3 seem to exhibit special kinetics to influenza infection and should be investigated further.
Genes of profile 3 play a key role in the mouse pneumonia model as determined by STC-GO analysis
GO analysis was applied to the genes belonging to profiles 1, 2, 3, 4, 7, 9, and 11. This method was employed to determine the main function of the genes that show the same expression trend according to GO. The key GOs identified in profiles 1 and 4 (Fig. S3A), profiles 2 and 7 (Fig. S3B), profiles 11 and 9 (Fig. S3C), and profile 3 (Fig. S3D) are displayed by bar graphs. Surprisingly, GOs related to PR8 virus-induced inflammatory and immune functions, including innate immune response, inflammatory response, immune response, cellular response to interferon-beta, were detected. The key GOs for profiles 1, 2, and in particular 3 were all closely related to PR8 virus-induced inflammatory and immune functions, with the exception of the apoptosis process. Whereas the key GOs belonging to profiles 4, 7, 9, and 11 were less relevant to inflammatory and immune functions.
This method of STC analysis explicitly took into account the dynamic nature of the temporal mRNA expression profiles during clustering and identified a number of distinct clusters. In this study, profile 3 was the only profile in which gene expression levels peaked on day 2 and then remained unchanged. Profile 3 also had the largest number of GOs and was most closely related to PR8 virus-induced inflammatory and immune pathways.
Genes of profile 3 were closely related to inflammation-related pathways in the mouse pneumonia model
Pathway analysis was performed to determine the predominant GOs of the genes in the seven significant clusters, and STC-pathway analysis was performed to filter the significant GOs of the genes in profile 3 (Fig. S4). The results indicated that the key pathways included in profile 3 were the NOD-like receptor signaling pathway, cytokine–cytokine receptor interactions, the Toll-like receptor signaling pathway, the chemokine signaling pathway, cell adhesion molecules (CAMs), the RIG-I-like receptor signaling pathway, and the Jak-STAT signaling pathway, all closely related to inflammatory and immune pathways.
Identification of the key genes belonging to profile 3 in the mouse pneumonia model by dynamic-gene network analysis
A dynamic-gene network was built according to the normalized signal intensity of specific genes. In studying some properties of the networks, k-cores in graph theory were introduced as a method of simplifying the analysis of graph topologies. A k-core of a network is a sub-network in which all nodes are connected to at least k other genes in the sub-network. A k-core of a protein–protein interaction network usually contains cohesive groups of proteins. In this study, genes in profiles 1, 2, 3, 4, 7, 9, and 11 were analyzed by a dynamic-gene network with a k-core algorithm (Fig. S5). The network reflects the correlations between genes. Each node describes a given gene, and the relationship between a pair of genes is represented by a line segment. Within the gene network, degree describes the number of links one gene has to others, with the most central genes in the network having the highest degree values. K-core in graph theory was applied to describe the characteristics of the network. A k-core of the dynamic-gene network usually contains cohesive groups of genes.
In our study, according to the genes in profile 3 (Table S2) and those constituting the dynamic-gene network (Table S3), it was clear that the main k-core (≥8) genes all belonged to profile 3, which included 63 genes, and the maximum k-core value was 15, for which there were 23 genes (Fig. S5).
According to STC-GO analysis of profile 3 (Fig. S3B), the five predominant GOs were identified as the innate immune response, defense response to virus, response to virus, inflammatory response, and immune response, and the 12 key genes (k-core = 15) related to these categories are listed in Table 1 (Fig. 3).
Key genes up-regulated during mouse pneumonia. The five predominant GOs represented among the genes were innate immune response, defense response to virus, response to virus, inflammatory response, and immune response. The related genes (k-core = 15) were Dhx58, Cxcl10, Cxcl11, Zbp1, Ifit1, Ifih1, Trim25, Mx2, Oas2, Cd274, Irgm1, and Irf7
Expression of the key genes detected by real-time RT-PCR
Real-time RT-PCR was performed to verify the expression levels of the 12 key genes (Dhx58, Cxcl10, Cxcl11, Zbp1, Ifit1, Ifih1, Trim25, Mx2, Oas2, Cd274, Irgm1, and Irf7) in the mouse lungs (Fig. 4). The results were consistent with the microarray data. It was worth noting that Cxcl10 and Cxcl11 are chemokine ligands; Ifit1, Ifih1, and Irf7 are all related to interferon function; and Mx2 is a natural barrier to influenza virus replication in the host.
Comparison of microarray and real-time RT-PCR analyses of 12 differentially expressed genes. Real-time RT-PCR was performed to verify the expression levels of the 12 key genes (A-L: Dhx58, Cxcl10, Zbp1, Ifit1, Ifih1, Trim25, Mx2, Oas2, Cd274, Cxcl11, Irgm1, and Irf7)) in the mouse lungs. The results were consistent with the microarray data
Discussion
Influenza A virus infection is one of the major causes of disease-related mortality worldwide. H1N1, a highly pathogenic subtype of influenza A virus, causes acute respiratory infections, which in some cases leads to severe pneumonia and death. Signal transduction between genes displays the dynamic characteristics of gene interactions, and these interactions constitute complex dynamic regulatory networks. Analysis of these networks can reveal dynamic changes in the genome under various pathophysiological conditions. The stronger the signal for a differentially expressed gene, the more crucial the role of that gene in the dynamic-gene network and in pathogenesis of the virus. To date, the dynamic-gene network for severe pneumonia induced by H1N1 virus infection remained unreported, although microarray studies of gene expression in the influenza-infected mouse lung have been done, but they focus on genes related with key immune cells [22] or transcriptomics [23]. Different from previous studies, in this study, we focus on kinetics of interaction between genes; exploring dynamic changes in the genome and identifying the key genes regulated during pneumonia induced by H1N1 influenza virus may provide important insight into the pathogenesis of this virus.
In a previous study, we revealed 82 microRNAs and 3371 mRNAs were differentially expressed by gene chips. The 82 microRNAs were further analyzed with STC analysis, and 46 microRNAs were shown to be significantly expressed. And then using potential target genes of the 46 microRNAs, we looked for intersections of these 3371 differentially expressed mRNAs and 719 intersection genes were indentified. Focusing on the 46 microRNAs and 719 genes, we revealed microRNA expression profiles and networks in mouse lung infected with H1N1 influenza virus [24]. In this study, we focused on the 3371 differentially expressed mRNAs and aimed to reveal the key dynamic genes induced in expression by H1N1 influenza virus in a mouse pneumonia model. The RVM and F-test were then applied to filter the differentially expressed genes. Seven gene clusters were confirmed to be significantly altered in expression; these were profiles 1, 2, 3, 4, 7, 9, and 11. It is worth noting that the opposite trend was observed between profiles 1 and 4, profiles 2 and 7, and profiles 11 and 9. In profile 3, gene expression was up-regulated, with expression levels peaking at day 2, then remaining constant. STC-GO analysis revealed GOs related to H1N1 virus-induced inflammatory and immune pathways, including innate immune response, inflammatory response, immune response, and cellular response to interferon-beta. When focusing on profiles 1, 2, and 3, the key GOs for profile 3 were all closely related to H1N1 virus-induced inflammatory and immune pathways, with the exception of the apoptosis process. STC-pathway analysis also revealed that the genes of profile 3 were closely associated with inflammation-related pathways, such as the NOD-like receptor signaling pathway, cytokine–cytokine receptor interactions, the Toll-like receptor signaling pathway, the chemokine signaling pathway, CAMs, the RIG-I-like receptor signaling pathway, and the Jak-STAT signaling pathway in the mouse pneumonia model. At the same time, STC-pathway analysis results showed the value of these pathways; according to the information of STC-pathway analysis, all above pathways have key value in a mouse pneumonia model, but the NOD-like receptor signaling pathway is the most important pathway and so on. The dynamic-gene network revealed that the key genes belonged to profile 3 and five predominant GOs (innate immune response, defense response to virus, response to virus, inflammatory response, and immune response) with maximum k-core values, and these genes were Dhx58, Cxcl10, Cxcl11, Zbp1, Ifit1, Ifih1, Trim25, Mx2, Oas2, Cd274, Irgm1, and Irf7. Cxcl10 and Cxcl11 were chemokine ligands; Ifit1, Ifih1, and Irf7 were all related to interferon function; and Mx2 was a natural barrier to influenza virus replication in the host. These findings will form the basis for future in-depth studies, particularly related to the 12 key genes closely associated with inflammation-related pathways.
In this study, it was not practical to include analysis of genes that were down-regulated in response to H1N1 influenza virus infection, but our further studies could explore the down-regulated genes which belong to profile 4 or profile 7 antagonized with profile 1 or profile 2. We believed that pairwise analysis of antagonistic gene clusters will be more meaningful and interesting due to more emphasis on functional verification.
In conclusion, our results suggested that changes in the expression of gene clusters and changes in the complex interactions between genes significantly altered gene functions during mouse pneumonia. Key genes have been identified that display dynamic gene expression and mediate important pathogenic functions. These results are a prelude to advancements in mouse H1N1 influenza virus infection biology, and aid the use of mice as a model organism for human H1N1 influenza virus infection studies.
References
K. Stöhr, Lancet Infect. Dis. 2(9), 517 (2002)
D. Butler, Nature 464(7292), 1112–1113 (2010)
G. Neumann, Y. Kawaoka, Virology 287(2), 243–250 (2001)
S. Sun, G. Zhao, W. Xiao, J. Hu, Y. Guo, H. Yu, X. Wu, Y. Tan, Y. Zhou, Virol. J. 8, 52 (2011)
G.W. Wright, R.M. Simon, Bioinformatics 19(18), 2448–2455 (2003)
H. Yang, N. Crawford, L. Lukes, R. Finney, M. Lancaster, K.W. Hunter, Clin. Exp. Metastasis 22(7), 593–603 (2005)
R. Clarke, H.W. Ressom, A. Wang, M.C. Liu, E.A. Gehan, Y. Wang, Nat. Rev. Cancer 8(1), 37–49 (2008)
S. Xiao, D. Mo, Q. Wang, J. Jia, L. Qin, X. Yu, Y. Niu, X. Zhao, X. Liu, Y. Chen, BMC Genom. 7(11), 544 (2010)
L.D. Miller, P.M. Long, L. Wong, S. Mukherjee, L.M. McShane, E.T. Liu, Cancer Cell 2(5), 353–361 (2002)
M.F. Ramoni, P. Sebastiani, I.S. Kohane, Proc. Natl. Acad. Sci. U.S.A. 99(14), 9121–9126 (2002)
Gene Ontology Consortium, Nucleic Acids Res. 34(Database issue), D322–D326 (2006)
M. Ashburner, C.A. Ball, J.A. Blake, D. Botstein, H. Butler, J.M. Cherry, A.P. Davis, K. Dolinski, S.S. Dwight, J.T. Eppig, M.A. Harris, D.P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J.C. Matese, J.E. Richardson, M. Ringwald, G.M. Rubin, G. Sherlock, Nat. Genet. 25(1), 25–29 (2000)
D. Dupuy, N. Bertin, C.A. Hidalgo, K. Venkatesan, D. Tu, D. Lee, J. Rosenberg, N. Svrzikapa, A. Blanc, A. Carnec, A.R. Carvunis, R. Pulak, J. Shingles, J. Reece-Hoyes, R. Hunt-Newbury, R. Viveiros, W.A. Mohler, M. Tasan, F.P. Roth, C. Le Peuch, I.A. Hope, R. Johnsen, D.G. Moerman, A.L. Barabasi, D. Baillie, M. Vidal, Nat. Biotechnol. 25(6), 663–668 (2007)
T. Schlitt, K. Palin, J. Rung, S. Dietmann, M. Lappe, E. Ukkonen, A. Brazma, Genome Res. 13(12), 2568–2576 (2003)
M. Kanehisa, S. Goto, S. Kawashima, Y. Okuno, M. Hattori, Nucleic Acids Res. 32(Database issue), D277–D280 (2004)
M. Yi, J.D. Horton, J.C. Cohen, H.H. Hobbs, R.M. Stephens, BMC Bioinform. 7, 30 (2006)
S. Draghici, P. Khatri, A.L. Tarca, K. Amin, A. Done, C. Voichita, C. Georgescu, R. Romero, Genome Res. 17(10), 1537–1545 (2007)
S. Xiao, J. Jia, D. Mo, Q. Wang, L. Qin, Z. He, X. Zhao, Y. Huang, A. Li, J. Yu, Y. Niu, X. Liu, Y. Chen, PLoS ONE 5(6), e11377 (2010)
M.R. Carlson, B. Zhang, Z. Fang, P.S. Mischel, S. Horvath, S.F. Nelson, BMC Genom. 7, 40 (2006)
C. Prieto, A. Risueno, C. Fontanillo, J. De las Rivas, PLoS ONE 3(12), e3911 (2008)
A.L. Barabási, Z.N. Oltvai, Nat. Rev. Genet. 5(2), 101–113 (2004)
J.C. Kash, T.M. Tumpey, S.C. Proll, V. Carter, O. Perwitasari, M.J. Thomas, C.F. Basler, P. Palese, J.K. Taubenberger, A. García-Sastre, D.E. Swayne, M.G. Katze, Nature 443(7111), 578–581 (2006)
F.X. Ivan, J.C. Rajapakse, R.E. Welsch, S.G. Rozen, T. Narasaraju, G.M. Xiong, B.P. Engelward, V.T. Chow, Funct. Integr. Genomics 12(1), 105–117 (2012)
Y. Bao, Y. Gao, Y. Jin, W. Cong, X. Pan, X. Cui, Mol. Genet. Genomics 290(5), 1885–1897 (2015)
Acknowledgements
This work is supported by the China Academy of Chinese Medical Sciences Foundation (No. ZZ2014010, No. ZZ2014040). The authors would also like to thank Li Jing from Gminix Company for technical assistance.
Author contributions
YYB, YJG, and YJS carried out the mouse experiments. YYB carried out microarray and bioinformatics analysis. YYB participated in the design of the study, performed the statistical analysis, and drafted the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interests.
Ethical approval
All procedures involving animals were approved by the Institute of Animal Care and Use Committee at the China Academy of Traditional Chinese Medicine. The animal experiments were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the China Academy of Traditional Chinese Medicine.
Additional information
Edited by William Dundon.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bao, Y., Gao, Y., Shi, Y. et al. Dynamic gene expression analysis in a H1N1 influenza virus mouse pneumonia model. Virus Genes 53, 357–366 (2017). https://doi.org/10.1007/s11262-017-1438-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-017-1438-y