Abstract
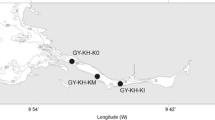
The applicability of microarrays to monitor harmful algae across a broad range of ecological niches and toxic species responsible for harmful algal events has been one of the key tasks in the EU Seventh Framework Programme (FP7)-funded Microarrays for the Detection of Toxic Algae project. The technique has a strong potential for improving speed and accuracy of the identification of harmful algae and their toxins to assist monitoring programmes. Water samples were collected from a number of coastal sites around Ireland, including several that are used in the Irish National Phytoplankton and Biotoxin Monitoring Programme. Ribosomal RNA was extracted from filtered field samples, labelled with a fluorescent dye, and hybridised to probes spotted in a microarray format on a glass slide. The fluorescent signal intensity of the hybridisation to >120 probes on the chip was analysed and compared with actual field counts. There was a general agreement between cell counts and microarray signal. Results are presented for field samples taken from a range of stations along the Irish coastline known for harmful algal events during the first field trial (July 2009–April 2010).






Similar content being viewed by others
References
Bowers HA, Tomas C, Tengs T, Kempton JW, Lewitus AJ, Oldach DW (2006) Raphidophyceae (Chadefaud Ex Silva) systematics and rapid identification: sequence analysis and real-time PCR assays. J Phycol 42:1333–1348
Browne R, Deegan B, O’Carroll T, Norman M, O’Cinneide M (2007) Status of Irish Aquaculture, 2006. Marine Institute/Bord Iascaigh Mhara/Taighide Mara Teo, pp 113
Campàs M, Prieto-Simón B, Marty JL (2007) Biosensors to detect marine toxins: assessing seafood safety. Talanta 72:884–895
Chen GF, Wang GC, Zhang CY, Zhang BY, Wang XK, Zhou BC (2008) Development of rRNA and rDNA-targeted probes for fluorescence in situ hybridization to detect Heterosigma akashiwo (Raphidophyceae). J Exp Mar Bio Ecol 355:66–75
Diercks S, Metfies K, Medlin LK (2008) Molecular probe sets for the detection of toxic algae for use in sandwich hybridization formats. J Plankton Res 30:439–448
Dittami SM, Edvardsen B (2012) GPR-analyzer: a simple tool for quantitative analysis of hierarchical multispecies microarrays. Env Sci Poll Res. doi:10.1007/s11356-012-1051-5
Eller G, Töbe K, Medlin LK (2007) Hierarchical probes at various taxonomic levels in the Haptophyta and a new division level probe for the Heterokonta. J Plankton Res 29:629–640
Eppley RW, Holmes RW, Strickland JDH (1967) Sinking rates of marine phytoplankton measured with a fluorometer. J Exp Mar Biol Ecol 1:191–208
Evans KM, Bates SS, Medlin LK, Hayes PK (2004) Microsatellite marker development and genetic variation in the toxic marine diatom Pseudo-nitzschia multiseries (Bacillariophyceae). J Phycol 40:911–920
Galluzzi L, Bertozzini E, Penna A, Perini F, Pigalarga A, Graneli E, Magnani M (2008) Detection and quantification of Prymnesium parvum (Haptophyceae) by real-time PCR. Appl Microbiology 46:261–266
GEOHAB (2001) GEOHAB Science Plan. Gilbert P, Pitcher. G (eds) SCOR and IOC: Baltimore. pp 87
Guillou L, Nezan E, Cueff V, Denn EEL, Cambon-Bonavita MA, Gentien P, Barbier G (2002) Genetic diversity and molecular detection of three toxic dinoflagellate genera (Alexandrium, Dinophysis and Karenia) from French coasts. Protist 153:223–238
Guschin DY, Mobarry BK, Proudnikov D, Stahl DA, Rittmann BE, Mirzabekov AD (1997) Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl Environ Microbiol 63:2397–2402
Hasle GR (1978) Diatoms in phytoplankton manual Sournia A. UNESCO, Paris, pp 136–142
John U et al (2003) The application of a molecular clock based on molecular sequences and the fossil record to explain biogeographic distributions within the Alexandrium tamarense species complex (Dinophyceae). Mol Biol Evol 20:1015–1027
Karlson B, Cusack C, Bresnan E (2010) Intergovernmental Oceanographic Commission of UNESCO. IOC Manuals and Guides no. 55, Paris, p 110
Kavanagh S, Brennan C, O’Connor L, Moran S, Salas R, Lyons J, Silke J, Maher M (2010) Real-time PCR detection of Dinophysis species in Irish coastal waters. Mar Biotechnol 12:534–542
Lange M, Guillou L, Vaulot D, Simon N, Amann RI, Ludwig W, Medlin LK (1996) Identification of the Class Prymnesiophyceae and the genus Phaeocystis with rRNA-targeted nucleic acid probes. J Phycol 32:858–868
Lewis J, Medlin LK, Raine R (2012) MIDTAL (Microarrays for the Detection of Toxic Algae): a protocol for a successful microarray hybridisation and analysis. pp 96
Lilly EL, Kulis DM, Gentien P, Anderson DM (2002) Paralytic shellfish poisoning toxins in France linked to a human-introduced strain of Alexandrium catenella from the western Pacific: evidence from DNA and toxin analysis. J Plankton Res 24:443–452
Lim EL, Amaral LA, Caron DA, DeLong EF (1993) Application of rRNA-based probes for observing marine nanoplanktonic protists. Appl Environ Microbiol 59:1647–1655
Masseret E, Grzebyk D, Nagai S, Genovesi B, Lasserre B, Laabir M, Collos Y, Vaquer A, Berrebi P (2009) Unexpected genetic diversity among and within populations of the toxic Dinoflagellate Alexandrium catenella as revealed by nuclear microsatellite markers. Appl Environ Microbiol 75:2037–2045
McDermott G, Raine R (2010) Settlement bottle method for quantificative phytoplankton analysis. In: Karlson B, Cusack C, Bresnan E (eds) Microscopic and molecular methods for quantitative phytoplankton analysis. IOC of UNESCO, Paris, pp 21–24
Metfies K, Medlin LK (2008) Feasibility of transferring FISH-probes to an 18S rRNA genephylochip and a mapping of signal intensity. Appl Environ Microbiol 74:2814–2821
Metfies K, Huljic S, Lange M, Medlin LK (2005) Electrochemical detection of the toxic dinoflagellate Alexandrium ostenfeldii with a DNA-biosensor. Biosens Bioelectron 20:1349–1357
Mikulski CM, Morton SL, Doucette GJ (2005) Development and application of LSU rRNA probes for Karenia brevis in the Gulf of mexico, USA. Harmful Algae 4:49–60
Miller PE, Scholin CA (1998) Identification and enumeration of cultured and wild Pseudo-nitzschia (Bacillariophyceae) using species-specific LSU rRNA-targeted fluorescent probes and filter-based whole cell hybridization. J Phycol 34:371–382
Moon-van der Staay SY, De Wachter R, Vaulot D (2001) Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607–610
Penna A, Bertozzini E, Battocchi C, Galluzzi L, Giacobbe MG, Vila M, Garces E, Luglie A, Magnani M (2007) Monitoring of HAB species in the Mediterranean Sea through molecular methods. J Plankton Res 29:19–38
Raine R, O’Boyle S, O’Higgins T, White M, Patching J, Cahill B, McMahon T (2001) A satellite and field portrait of a Karenia mikimotoi bloom off the south coast of Ireland, August, 1998. Hydrobiologia 465:187–193
Raine R, McDermott G, Silke J, Lyons K, Nolan G, Cusack C (2010) A simple short range model for the prediction of harmful algal events in the bays of southwestern Ireland. J Mar Sys 83:150–157
Scholin CA and Anderson DM (1998) Detection and quantification of HAB species using antibody and DNA probes: progress to date and future research objectives. In: Regura, B., Blanko, J., Fernandez, M.L. and Wyatt, T (eds). Harmful algae. Xunta de Galicia and Intergovernmental Ocenaographic Commission of UNESCO. pp. 253–257
Scholin CA, Miller P, Buck K, Chavez F, Harris P, Haydock P, Howard J, Cangelosi G (1997) Detection and quantification of Pseudo-nitzschia australis in cultured and natural populations using LSU rRNA-targeted probes. Limnol Oceanogr 42:1265–72
Silke J, O’Beirn F, Cronin M (2005) Karenia mikimotoi: an exceptional dinoflagellate bloom in western Irish waters, Summer 2005. In: Marine Environment and Health Series 21. Marine Institute, Galway, Ireland
Simon N, Brenner J, Edvardsen B, Medlin LK (1997) The Identification of Chrysochromulina and Prymnesium species (Haptophyta, Prymnesiophyceae) using fluorescent or chemiluminescent oligonucleotide probes: a means for improving studies on toxic algae. Eur J Phycol 32:393–401
Simon N, Campbell L, Örnólfsdóttir E, Groben R, Guillou L, Lange M, Medlin LK (2000) Oligonucleotide probes for the identification of three algal groups by dot blot and fluorescent whole-cell hybridization. J Eukaryot Microbiol 47:76–84
Throndsen J (1978) Preservation and storage. In: Sournia A (ed) Phytoplankton manual. UNESCO, Paris, pp 69–74
Töbe K, Eller G, Medlin LK (2006) Automated detection and enumeration for toxic algae by solid-phase cytometry and the introduction of a new probe for Prymnesium parvum (Haptophyta: prymnesiophyceae). J Plankton Res 28:643–657
Touzet N, Franco JM, Raine R (2007) Characterisation of non-toxic and toxin producing populations of Alexandrium minutum (Dinophyceae) in Irish coastal waters. Appl Environ Microbiol 73:3333–3342
Touzet N, Keady E, Raine R, Maher M (2009) Evaluation of taxa-specific real-time PCR, whole cell FISH and morphotaxonomy analyses for the detection and quantification of the toxic microalgae Alexandrium minutum (Dinophyceae), Global Clade ribotype. FEMS Microbiol Ecol 67:329–341
Touzet N, Davidson K, Pete R, Flanagan K, McCoy GR, Amzil Z, Maher M, Chapelle A, Raine R (2010) Co-occurrence of the West European (Gr. III) and North American (Gr. I) ribotypes of Alexandrium tamarense (Dinophyceae) in Shetland, Scotland. Protist 161:370–384
Touzet N, Lacaze JP, Maher M, Turrel E, Raine R (2011) Summer dynamics of Alexandrium ostenfeldii (Dinophyceae) and spirolide toxins in Cork Harbour, Ireland. Mar Ecol Prog Ser 425:21–33
Tyrrell JV, Bergquist PR, Bergquist PL, Scholin CA (2001) Detection and enumeration of Heterosigma akashiwo and Fibrocapsa japonica (Raphidophyceae) using rRNA-targeted oligonucleotide probes. Phycol 40:457–467
Vilariño N, Fonfría ES, Louzao C, Botana M (2009) Use of biosensors as alternatives to current regulatory methods for marine biotoxins. Sensors 9:9414–9443
Acknowledgments
This work was funded through the EU 7th Framework Programme (FP7-ENV-2007-1-MIDTAL-201724). The authors would like to acknowledge the assistance of the captain and crew of R.V Celtic Explorer. Special thanks to Donal Geary of the r/v John Boy and to colleagues Annette Wilson, Evelyn Keady, Hazel Farrell, and Sarah Cosgrove from NUI Galway for their assistance in field studies. The scientific and technical assistance of Linda Medlin and co-workers of the MIDTAL project team is also greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Rights and permissions
About this article
Cite this article
McCoy, G.R., Touzet, N., Fleming, G.T. et al. An evaluation of the applicability of microarrays for monitoring toxic algae in Irish coastal waters. Environ Sci Pollut Res 20, 6751–6764 (2013). https://doi.org/10.1007/s11356-012-1294-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-012-1294-1