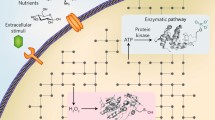
Abstract
Biological macromolecules (proteins, nucleic acids, polysaccharides, etc.) are the building blocks of life, which constantly undergo chemical modifications that are often reversible and spatial-temporally regulated. These dynamic properties of chemical modifications play fundamental roles in physiological processes as well as pathological changes of living systems. The Major Research Project (MRP) funded by the National Natural Science Foundation of China (NSFC)—“Dynamic modifications of biomacromolecules: mechanism and chemical interventions” aims to integrate cross-disciplinary approaches at the interface of chemistry, life sciences, medicine, mathematics, material science and information science with the following goals: (i) developing specific labeling techniques and detection methods for dynamic chemical modifications of biomacromolecules, (ii) analyzing the molecular mechanisms and functional relationships of dynamic chemical modifications of biomacromolecules, and (iii) exploring biomacromolecules and small molecule probes as potential drug targets and lead compounds.
Similar content being viewed by others
References
Aguzzi, A., and Altmeyer, M. (2016). Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol 26, 547–558.
Ai, H., Guo, Y., Sun, D., Liu, S., Qi, Y., Guo, J., Qu, Q., Gong, Q., Zhao, S., Li, J., et al. (2019). Examination of the deubiquitylation site selectivity of USP51 by using chemically synthesized ubiquitylated histones. Chembiochem 20, 221–229.
Arrowsmith, C.H., Audia, J.E., Austin, C., Baell, J., Bennett, J., Blagg, J., Bountra, C., Brennan, P.E., Brown, P.J., Bunnage, M.E., et al. (2015). The promise and peril of chemical probes. Nat Chem Biol 11, 536–541.
Bandeira, N., Tsur, D., Frank, A., and Pevzner, P.A. (2007). Protein identification by spectral networks analysis. Proc Natl Acad Sci USA 104, 6140–6145.
Black, J.C., Van Rechem, C., and Whetstine, J.R. (2012). Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 48, 491–507.
Bode, A.M., and Dong, Z. (2004). Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 4, 793–805.
Bunnage, M.E., Chekler, E.L.P., and Jones, L.H. (2013). Target validation using chemical probes. Nat Chem Biol 9, 195–199.
Bunnage, M.E., Gilbert, A.M., Jones, L.H., and Hett, E.C. (2015). Know your target, know your molecule. Nat Chem Biol 11, 368–372.
Casey, P.J. (1995). Protein lipidation in cell signaling. Science 268, 221–225.
Cedar, H., and Bergman, Y. (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10, 295–304.
Chatterjee, J., and Köhn, M. (2013). Targeting the untargetable: recent advances in the selective chemical modulation of protein phosphatase-1 activity. Curr Opin Chem Biol 17, 361–368.
Chu, G.C., Pan, M., Li, J., Liu, S., Zuo, C., Tong, Z.B., Bai, J.S., Gong, Q., Ai, H., Fan, J., et al. (2019). Cysteine-aminoethylation-assisted chemical ubiquitination of recombinant histones. J Am Chem Soc 141, 3654–3663.
Cohen, P. (2002). The origins of protein phosphorylation. Nat Cell Biol 4, E127–E130.
Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823.
Crick, F. (1970). Central dogma of molecular biology. Nature 227, 561–563.
Davis, B.G. (2004). Mimicking posttranslational modifications of proteins. Science 303, 480–482.
Dawson, M.A., Kouzarides, T., and Huntly, B.J.P. (2012). Targeting epigenetic readers in cancer. N Engl J Med 367, 647–657.
Goldberg, A.D., Allis, C.D., and Bernstein, E. (2001). Epigenetics: a landscape takes shape. Cell 128, 635–638.
Goll, M.G., and Bestor, T.H. (2005). Eukaryotic cytosine methyl transferases. Annu Rev Biochem 74, 481–514.
Greer, P.L., Hanayama, R., Bloodgood, B.L., Mardinly, A.R., Lipton, D.M., Flavell, S.W., Kim, T.K., Griffith, E.C., Waldon, Z., Maehr, R., et al. (2010). The Angelman syndrome protein Ube3A regulates synapse development by ubiquitinating Arc. Cell 140, 704–716.
Gregorich, Z.R., and Ge, Y. (2014). Top-down proteomics in health and disease: challenges and opportunities. Proteomics 14, 1195–1210.
Hang, H.C., and Linder, M.E. (2011). Exploring protein lipidation with chemical biology. Chem Rev 111, 6341–6358.
Heim, C., and Binder, E.B. (2012). Current research trends in early life stress and depression: review of human studies on sensitive periods, gene-environment interactions, and epigenetics. Exp Neurol 233, 102–111.
Holoch, D., and Moazed, D. (2015). RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16, 71–84.
Ibraheem, A., and Campbell, R.E. (2010). Designs and applications of fluorescent protein-based biosensors. Curr Opin Chem Biol 14, 30–36.
Janke, C., and Chloë Bulinski, J. (2011). Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol 12, 773–786.
Jones, P.A. (2002). DNA methylation and cancer. Oncogene 21, 5358–5360.
Kochendoerfer, G.G., and Kent, S.B. (1999). Chemical protein synthesis. Curr Opin Chem Biol 3, 665–671.
Li, J., and Chen, P.R. (2016). Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat Chem Biol 12, 129–137.
Li, J., Kong, H., Huang, L., Cheng, B., Qin, K., Zheng, M., Yan, Z., and Zhang, Y. (2018). Visible light-initiated bioorthogonal photoclick cycloaddition. J Am Chem Soc 140, 14542–14546.
Lin, W., Gao, L., and Chen, X. (2015). Protein-specific imaging of posttranslational modifications. Curr Opin Chem Biol 28, 156–163.
Luo, G.Z., Blanco, M.A., Greer, E.L., He, C., and Shi, Y. (2015). DNA N6-methyladenine: a new epigenetic mark in eukaryotes? Nat Rev Mol Cell Biol 16, 705–710.
Maddika, S., and Chen, J. (2009). Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat Cell Biol 11, 409–419.
Magi, B., Bargagli, E., Bini, L., and Rottoli, P. (2006). Proteome analysis of bronchoalveolar lavage in lung diseases. Proteomics 6, 6354–6369.
Mann, M., and Jensen, O.N. (2003). Proteomic analysis of post-translational modifications. Nat Biotechnol 21, 255–261.
Moremen, K.W., Tiemeyer, M., and Nairn, A.V. (2012). Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol 13, 448–462.
Ohtsubo, K., and Marth, J.D. (2006). Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867.
Ooi, S.K.T., and Bestor, T.H. (2008). The colorful history of active DNA demethylation. Cell 133, 1145–1148.
Pan, M., Zheng, Q., Ding, S., Zhang, L., Qu, Q., Wang, T., Hong, D., Ren, Y., Liang, L., Chen, C., et al. (2019). Chemical protein synthesis enabled mechanistic studies on the molecular recognition of K21-linked ubiquitin chains. Angew Chem Int Ed 58, 2627–2631.
Pettitt, J., Zeitlin, L., Kim, D.H., Working, C., Johnson, J.C., Bohorov, O., Bratcher, B., Hiatt, E., Hume, S.D., Johnson, A.K., et al. (2013). Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci Transl Med 5, 199ra113.
Prabakaran, S., Lippens, G., Steen, H., and Gunawardena, J. (2012). Post-translational modification: nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip Rev Syst Biol Med 4, 565–583.
Radivojac, P., Vacic, V., Haynes, C., Cocklin, R.R., Mohan, A., Heyen, J. W., Goebl, M.G., and Iakoucheva, L.M. (2010). Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78, 365–380.
Ramil, C.P., and Lin, Q. (2014). Photoclick chemistry: a fluorogenic light-triggered in vivo ligation reaction. Curr Opin Chem Biol 21, 89–95.
Rix, U., and Superti-Furga, G. (2008). Target profiling of small molecules by chemical proteomics. Nat Chem Biol 5, 616–624.
Ross, C.A., and Poirier, M.A. (2004). Protein aggregation and neurodegenerative disease. Nat Med 10, S10–S17.
Roundtree, I.A., and He, C. (2016). Nuclear m6A reader YTHDC1 regulates mRNA splicing. Trends Genet 32, 320–321.
Rubin, C.S., and Rosen, O.M. (1975). Protein phosphorylation. Annu Rev Biochem 44, 831–887.
Schenone, M., Dančik, V., Wagner, B.K., and Clemons, P.A. (2013). Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol 9, 232–240.
Snider, N.T., and Omary, M.B. (2014). Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nat Rev Mol Cell Biol 15, 163–177.
Taira, N., Nihira, K., Yamaguchi, T., Miki, Y., and Yoshida, K. (2007). DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell 25, 725–738.
Walsh, C.T., Garneau-Tsodikova, S., and Gatto, G.J. (2005). Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed 44, 7342–7372.
Wang, J., Liu, Y., Liu, Y., Zheng, S., Wang, X., Zhao, J., Yang, F., Zhang, G., Wang, C., and Chen, P.R. (2019). Time-resolved protein activation by proximal decaging in living systems. Nature 569, 509–513.
Wells, L., Vosseller, K., and Hart, G.W. (2001). Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science 291, 2376–2378.
Wold, F. (1981). In vivo chemical modification of proteins (posttranslational modification). Annu Rev Biochem 50, 783–814.
Xiao, X., Tang, J.J., Peng, C., Wang, Y., Fu, L., Qiu, Z.P., Xiong, Y., Yang, L.F., Cui, H.W., He, X.L., et al. (2011). Cholesterol modification of Smoothened is required for hedgehog signaling. Mol Cell 66, 154–162. 10.
Zentner, G.E., and Henikoff, S. (2013). Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol 20, 259–266.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supporting Information
Table S1 List of research projects that have been funded by the Major Research Project at NSFC
The supporting information is available online at http://life.scichina.com and https://link.springer.com. The supporting materials are published as submitted, without typesetting or editing. The responsibility for scientific accuracy and content remains entirely with the authors.
Supporting Information
Rights and permissions
About this article
Cite this article
Wang, C., Zou, P., Yang, C. et al. Dynamic modifications of biomacromolecules: mechanism and chemical interventions. Sci. China Life Sci. 62, 1459–1471 (2019). https://doi.org/10.1007/s11427-019-9823-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-019-9823-1